xGen™ DNA Library Preparation Kits
Comprehensive coverage with fewer PCR duplicates
Streamline preparation of comprehensive NGS libraries from dsDNA. These DNA library preparation kits utilize enzymatic fragmentation (EZ) or mechanical shearing (MC) methods to generate libraries suitable for PCR-free, PCR-amplified, and targeted sequencing research applications.
xGen NGS—made for DNA library preparation.
xGen DNA Library Prep Kit EZ and EZ UNI
- Low input, high complexity—up to 3x fewer duplicates from 1 ng of DNA.
- Improved yield for enrichment—specially formulated PCR master mix boosts yield from a minimal number of PCR cycles.
- High multiplex capability—pre-plated UDI primers enable multiplexing of up to 1536 samples.
- Compatible with xGen Normalase™ Module—streamlines normalization of sequencing libraries from multiplexing
- Workflow design for easy automation
- Skip the conversion step. Make native libraries for DNBSEQ™, Element Biosciences AVITI™, and Ultima Genomics UG100™.
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
Ordering
Product details
Low input and PCR-free workflows
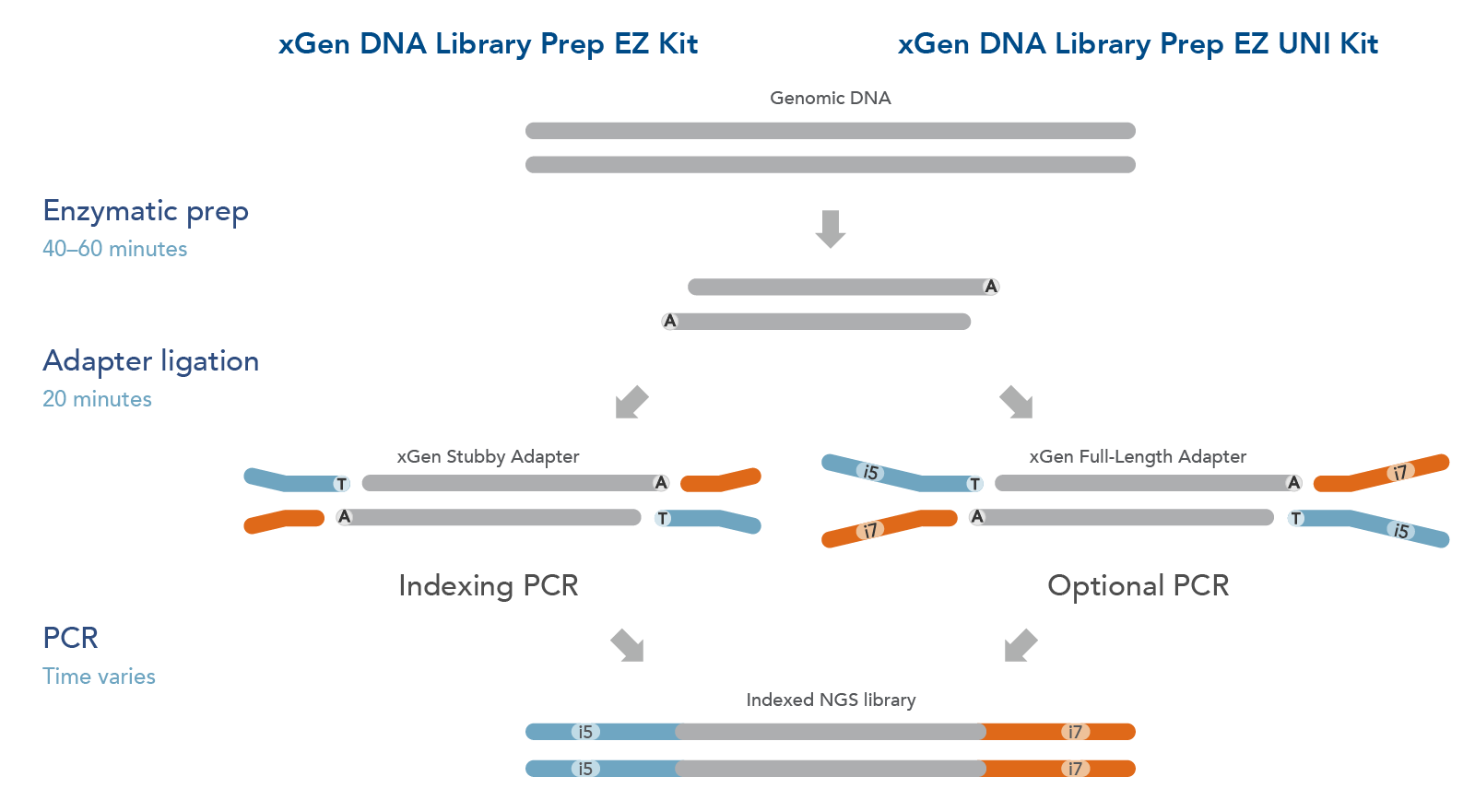
The xGen DNA Library EZ Kits are available in two configurations (Figure 1): xGen DNA Library EZ kit (Standard kit) and xGen DNA Library EZ UNI kit (Universal kit).
- The xGen DNA Library EZ kit supports indexing by PCR workflow, it includes xGen Stubby Adapters. Indexing primers are supplied separately.
- The xGen DNA Library EZ UNI kit supports an indexing by ligation workflow for optional PCR amplification or PCR-free workflow, amplification primers are included in the kit. Indexed adapters are supplied separately.
Both DNA library preparation kits include an enzymatic fragmentation module, a high efficiency ligase, and a PCR Master Mix that can generate library yields sufficient for hybridization-based enrichment or direct sequencing. Both kits have workflows compatible with high throughput applications and have been automated on multiple liquid handlers.
Figure 1. Workflow for the xGen DNA Library Prep EZ Kits. xGen DNA Library Prep EZ Kits are compatible with either an indexing by PCR workflow using xGen Stubby Adapters included in the kit and indexing primers supplied separately (left) or an indexing by ligation workflow using full length, indexed Y adapters (right). Both kits include amplification reagents for Indexing PCR or optional PCR amplification.
Table 1. Supported applications
| Research applications | xGen DNA Library EZ Kit and EZ UNI Kit |
|---|---|
| Whole genome sequencing | ✓ |
| Whole exome sequencing | ✓ |
| Variant detection | Germline + somatic |
| Genotyping | ✓ |
| CNV detection | ✓ |
Table 2. Specifications
| Features | xGen DNA Library EZ Kit | xGen DNA Library EZ UNI Kit |
|---|---|---|
| Sample types | Fresh frozen tissue, genomic DNA, PCR amplicons, high quality FFPE* | |
| Range of input concentrations | 0.1–1000 ng | 0.1–1000 ng |
| Indexing compatibility | CDI primers up to 96-plex, compatible with xGen Normalase Module UDI primers up to 1536-plex, compatible with xGen Normalase Module | xGen indexing primers for custom, full-length and truncated adapters, compatible with xGen Normalase Module |
| System compatibility and multiplexing format | Illumina® sequencing instruments, Element Biosciences AVITI, DNBSEQ devices compatibility | Manual and automated. Compatibility with Element Biosciences AVITI, DNBSEQ devices, Ultima Genomics UG100. Please inquire for a list of compatible liquid handling robots and scripts. |
*Optimization of the enzymatic fragmentation step may be required.
xGen Deceleration Module enables additional control over fragment length with the xGen DNA Library EZ Kits.
The xGen Deceleration Module
- generates an average library insert size of 550 bp
- offers flexible fragmentation times on automation platforms
Comprehensive coverage
The xGen DNA Library EZ Kit was benchmarked against another supplier of DNA library prep kit that uses enzymatic fragmentation and a widely used HiFi DNA polymerase. Two different libraries for each concentration of input DNA (Coriell NA 12878; 100 ng, 10 ng, and 1 ng) with both kits were generated and evaluated for enrichment using the xGen Pan-Cancer Panel to assess library quality metrics (Table 3). For comparative analysis, identical SPRI®-based size selections (Beckman Coulter) were used for both library prep kits. Libraries were sequenced 2x101 bp on an Illumina® MiSeq® System using v2 chemistry, and reads were normalized to 460k per sample.
The xGen DNA Library EZ Kit required a lower number of PCR cycles to generate yields equivalent to the kit from the alternate supplier. At lower inputs, particularly at 1 ng, the xGen DNA Library EZ Kit provided higher target coverage, a significantly lower percentage of PCR duplicates, and better coverage uniformity compared to the other supplier’s kit. The xGen DNA Library EZ Kit also produced a more uniform insert size across the input quantities evaluated, when compared to the other kit.
Table 3. Targeted sequencing metrics.
| Library prep | Input (ng) | Yield ng/µl (PCR cycles) | Aligned insert (bp) | Duplicates (%) | Mean coverage (%) | 20X Coverage (%) | 30X Coverage (%) | 40X Coverage (%) | 50X Coverage (%) |
|---|---|---|---|---|---|---|---|---|---|
| xGen DNA Library EZ-sample 1 | 100 | 97 (5) | 235 | 0.78 | 49.1 | 97.3 | 89.4 | 70.9 | 45.9 |
| xGen DNA Library EZ- sample 2 | 95 (5) | 233 | 0.51 | 42.7 | 96.0 | 83.6 | 57.6 | 29.1 | |
| Other supplier, sample 1 | 78 (5) | 211 | 0.35 | 48.5 | 97.2 | 89.5 | 70.0 | 44.1 | |
| Other supplier, sample 2 | 84 (5) | 212 | 0.28 | 41.5 | 95.9 | 82.3 | 54.2 | 25.4 | |
| xGen DNA Library EZ-sample 1 | 10 | 68 (8) | 226 | 1.50 | 48.8 | 97.4 | 89.4 | 69.9 | 44.6 |
| xGen DNA Library EZ-sample 2 | 73 (8) | 223 | 1.17 | 42.5 | 96.0 | 83.0 | 56.2 | 28.7 | |
| Other supplier, sample 1 | 98 (11) | 250 | 1.61 | 47.8 | 96.3 | 86.5 | 66.7 | 42.2 | |
| Other supplier, sample 2 | 91 (11) | 253 | 1.23 | 41.9 | 94.5 | 79.0 | 53.1 | 28.3 | |
| xGen DNA Library EZ-sample 1 | 1 | 78 (11) | 229 | 8.8 | 42.1 | 96.2 | 82.5 | 54.7 | 27.3 |
| xGen DNA Library EZ-sample 2 | 77 (11) | 227 | 6.8 | 37.1 | 93.9 | 73.7 | 39.7 | 13.9 | |
| Other supplier, sample 1 | 100 (17) | 316 | 46.6 | 13.9 | 14.3 | 0.98 | 0.11 | 0.08 | |
| Other supplier, sample 2 | 103 (17) | 315 | 41.5 | 12.5 | 8.89 | 0.42 | 0.09 | 0.06 |
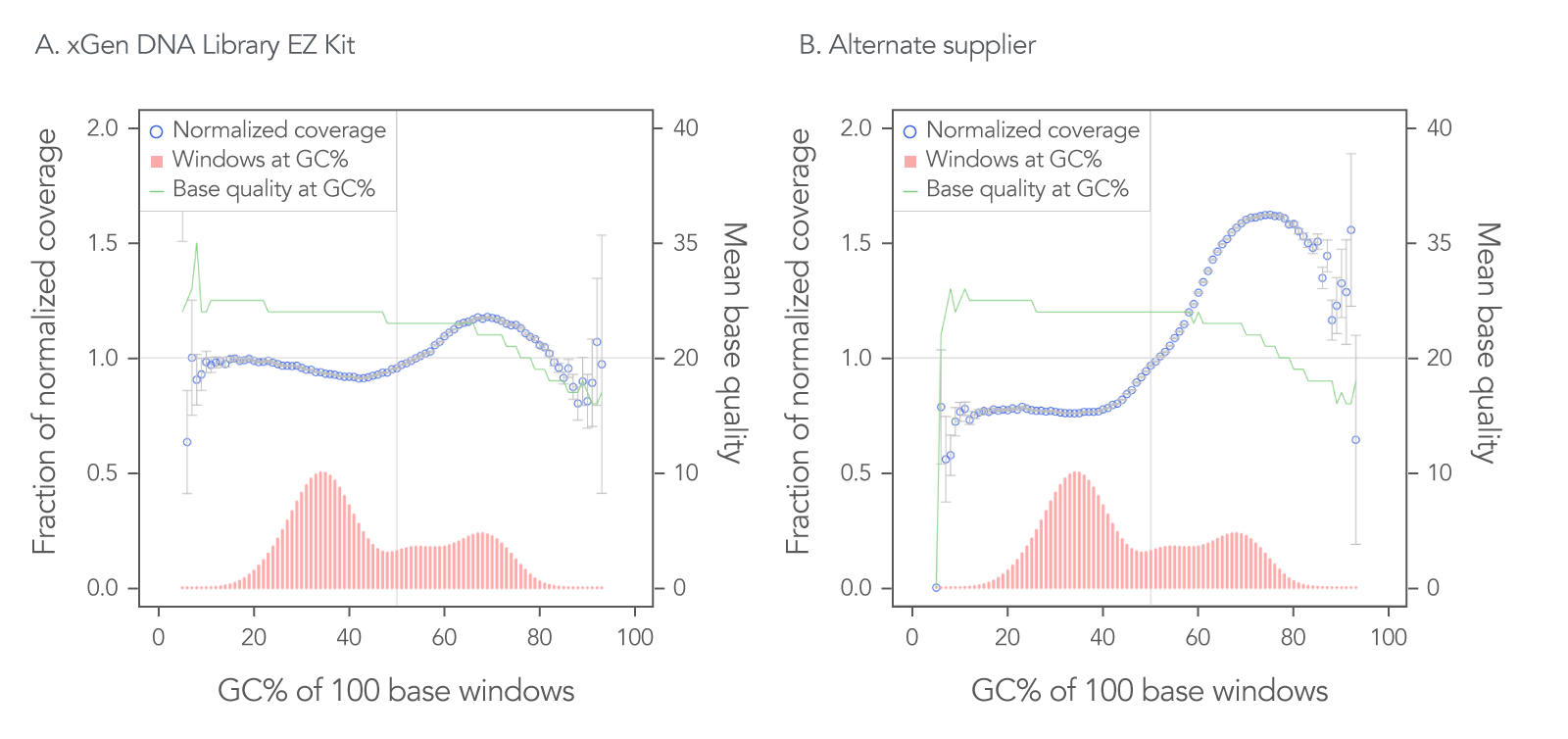
The xGen DNA Library EZ Kit was also benchmarked against another supplier of a DNA library kit using enzymatic fragmentation and 1 ng of DNA harvested from a mock bacterial community (ATCC® MSA-1000®). The DNA was converted into libraries using either the xGen DNA Library EZ Kit or the other enzymatic fragmentation library prep kit. The xGen DNA Library EZ Kit provided a greater library yield from fewer PCR cycles (Table 4), better normalized coverage across distinct GC compositions (Figure 2), improved coverage uniformity (Figure 2), and fewer PCR duplicates (Table 4) compared to the other supplier’s kit.
Figure 2. Higher normalized coverage observed using xGen DNA Library Prep EZ Kit versus another supplier of an enzymatic fragmentation-based library prep kit. To generate the two libraries, the xGen DNA Library EZ Kit (A) and a comparable, enzymatic fragmentation-based library prep kit from another supplier (B) was used on a mock bacterial community (ATCC, MSA-1000) with 1 ng DNA input. For comparative analysis, a 0.65x SPRI-based, DNA cleanup was used after PCR for both kits. The two libraries were sequenced 2x151 bp on an Illumina® MiniSeq® System in High Output mode. Reads were normalized to 5 M per sample. The mock bacterial community contained the following strains: B. cereus (GC% = 35.5), B. adolescentis (GC% = 59.4), C. beijerinckii (GC% = 29.9), D. radiodurans (GC% = 66.7), E. faecalis (GC% = 37.8), E. coli (GC% = 50.8), L. gasseri (GC% = 35.3), R. sphaeroides(GC% = 68.9), S. epidermidis (GC% = 32.0), and S. mutans (GC% = 36.8).
Table 4. Targeted sequencing metrics.
| Library prep kit | Library yield ng/µ L (PCR cycles) | Aligned insert (bp) | Duplicates (%) | Mean coverage (%) | 10X coverage (%) | 20X coverage (%) | 30X coverage (%) |
|---|---|---|---|---|---|---|---|
| xGen DNA Library EZ-MSA-sample 1 | 26 (11) | 313.7 | 0.69 | 33.4 | 98.1 | 95.1 | 64.7 |
| xGen DNA Library EZ-MSA-sample 2 | 26 (11) | 319.5 | 0.71 | 33.7 | 98.1 | 95.3 | 65.9 |
| Other supplier, sample-MSA-sample 1 | 4.4 (13) | 321.3 | 2.09 | 32.6 | 98.0 | 86.7 | 48.5 |
| Other supplier, sample-MSA-sample 2 | 4.7 (13) | 328 | 2.29 | 32.8 | 98.0 | 87.0 | 48.6 |
xGen DNA Library Prep MC Kit and MC UNI
- The xGen DNA Library Prep MC Kit is compatible with Covaris® sheared DNA—this kit supports 1 ng to 1 µg, PCR-free libraries from 50 ng input DNA.
- High multiplex capability—pre-plated UDI primers enable multiplexing of up to 1536 samples.
- Compatible with xGen Normalase™ Module—streamlines normalization of sequencing libraries from multiplexing
- Workflow design for easy automation
- Skip the conversion step. Make native libraries for DNBSEQ™, Element Biosciences AVITI™, and Ultima Genomics UG100™.
Product details
The xGen DNA Library Prep MC Kits offer a versatile solution to streamline dsDNA, NGS sample preparation for Illumina® sequencing platforms. This workflow processes fragmented DNA, such as Covaris®-sheared DNA, for rapid and efficient end repair, adenylation, and adapter ligation. It includes an optional library amplification step, thus enabling both manual and automated workflows. These DNA library preparation kits can also be used for targeted hybridization capture. The HiFi Polymerase Master Mix, which is supplied in the kit, is suitable for pre-hyb PCR to produce yields of 500 ng or more from as little as 1 ng DNA input. These kits are also compatible with any of the predesigned xGen Hyb Panels or a xGen Custom Hyb Panel.
Table 5. Specifications
| Feature | xGen DNA Library MC Kit | xGen DNA Library MC UNI Kit |
|---|---|---|
| Sample types | Genomic DNA extracted from tissue, blood, microbial isolates, and environmental samples | |
| Input concentration and type | 1 ng–1 µg input DNA in 50 µL volume | |
| Indexing compatibility | CDI primers up to 96-plex and UDI primers up to 1536-plex | xGen indexing primers for custom full-length and truncated adapters |
| System compatibility and multiplexing format | Illumina® sequencing instruments, Element Biosciences AVITI, DNBSEQ devices | Illumina sequencing instruments, Element Biosciences AVITI, DNBSEQ devices, Ultima Genomics UG100 |
| Workflow compatibility | Manual and automated. Please inquire for a list of compatible liquid handling robots and scripts. | |
Rapid, flexible workflow
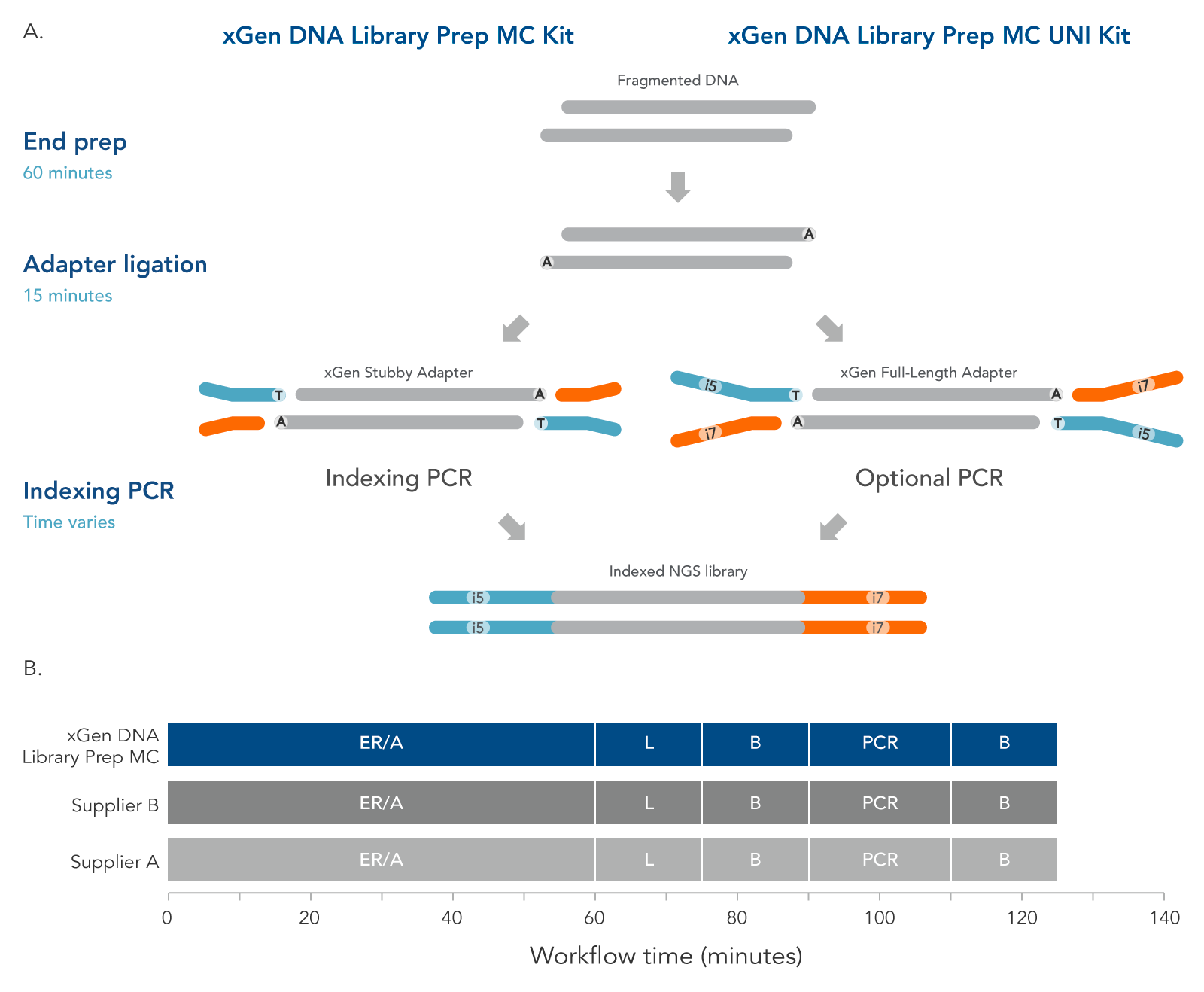
The xGen DNA Library Prep MC protocol (Figure 4) contains two enzymatic incubations, an indexing or optional PCR, and bead-purification steps. The protocol minimizes sample handling, shortening the overall, library preparation time to two hours with PCR.
Figure 3. Workflow for the xGen DNA Library Prep MC Kits. These kits are available in two configurations to support two indexing workflows. The incubation steps consist of end repair, polishing of dsDNA, and A-tailing, all performed in a single End Prep reaction followed by ligation of either a stubby Y adapter (xGen DNA Library Prep MC kit, left) or full-length indexed Y adapter (xGen DNA Library Prep MC UNI Kit, right). The xGen DNA Library Prep MC workflow incorporates an indexing PCR step after adapter ligation to complete the adapter sequences. PCR is an optional step for the MC UNI Kit. xGen DNA Library Prep MC Kits are compatible with various indexing primers and full-length indexed Y adapters, including UDI and UMI adapters and up to 1536 UDI primer pairs. Both kits are compatible with the Normalase module. The xGen DNA Library Prep MC workflow is comparable to other kit suppliers, where a 60-minute end repair/A-tailing step (ER/A) is followed by a 15-minute adapter ligation step (L), a bead-based purification (B), an optional library amplification step (PCR), and a second, bead-based purification (B).
Product data
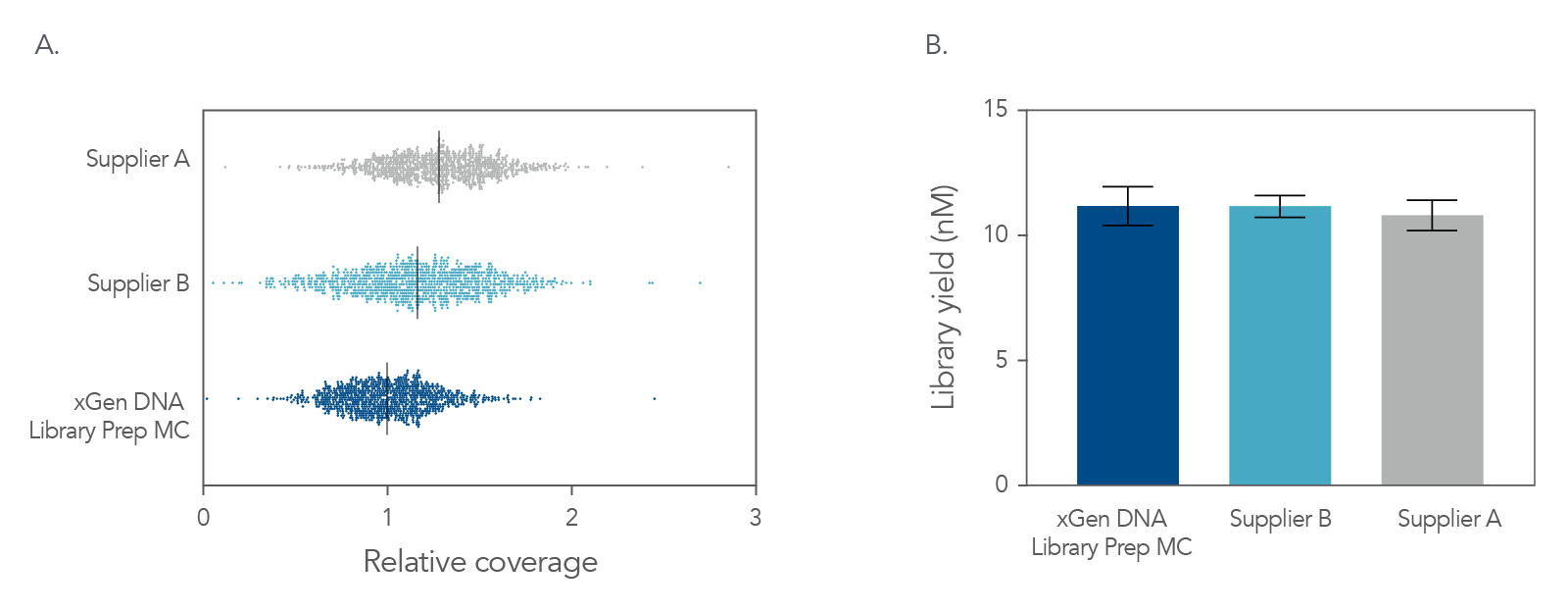
Figure 4. Balanced genome coverage. PCR-free libraries were constructed with the xGen DNA Library Prep MC UNI and two other suppliers’ kits, using 100 ng Coriell NA12878 DNA that was Covaris-sheared to 350 bp, in duplicate. All libraries were constructed using IDT for Illumina® TruSeq® UDI DNA Indexes (Cat. No. 20020590) at supplier-specified concentrations. A consistent bead ratio was used to generate equivalent insert sizes across supplier kits for direct comparison. Libraries were quantified by qPCR and sequenced on an Illumina® NovaSeq ® instrument, and data were normalized to 270 M reads per sample. Right Panel: PCR-free library yields were comparable across all three kits. Left Panel: xGen DNA Library Prep MC UNI showed a more balanced coverage of 971 high GC regions, relative to the mean coverage of other suppliers’ kits that included high GC regions greater than the mean coverage. A deviation from a relative coverage of 1 represents a reduction in coverage uniformity (GC rich bed file from Ross et al, Characterizing and Measuring Bias in Sequence Data; Genome Biol. 2013).
xGen Exome and Pan-Cancer Panels
The xGen DNA Library Prep MC and the two other suppliers’ kits were evaluated with the xGen Pan-Cancer and Exome enrichment panels using duplicate samples of Coriell NA12878 genomic DNA that was Covaris-sheared to 200 bp. A total of 10 ng sheared DNA was used for the xGen Pan-Cancer Panel, and 100 ng sheared DNA was used for the xGen Exome Panel v2. For Pan-Cancer, xGen DNA Library Prep MC and competitor libraries were prepared with truncated Y adapters at supplier-specified concentration and an xGen CDI Primer kit. The Exome libraries were prepared with the xGen DNA Library Prep MC UNI and other suppliers’ kits using IDT for Illumina® TruSeq® UD DNA Indexes (Cat. No. 20020590) at supplier-specified concentration. For both panels, a consistent bead ratio was used to generate equivalent insert sizes across supplier kits for direct comparison. Table 6 lists the experimental results.
The upper section of Table 6 lists data obtained in the following experiment: xGen Pan-Cancer captured libraries were sequenced on a MiSeq® with 100 bp, paired-end reads and were normalized to 1.5 M reads. xGen DNA Library Prep MC Kit yields using the HiFi Master Mix were equivalent to other supplier kits that used Kapa HiFi Hot Start Ready Mix, thus showing library amplification for pre-hyb PCR. Due to MiSeq chemistry, PCR duplicates directly reflect library complexity. All three kits demonstrated similar target coverage, estimated library size, and uniformity of target coverage.
The lower section of Table 6 lists data obtained in the following experiment: Exome capture libraries were sequenced on a NovaSeq® instrument (Illumina) with 150 bp, paired end reads and were normalized to 80 M reads. The xGen DNA Library Prep MC yields using the HiFi Master Mix were equivalent to other supplier kits that used Kapa HiFi Hot Start Ready Mix, thus showing comprehensive library amplification for pre-hyb PCR. Due to NovaSeq chemistry, PCR duplicates include cluster duplicates and do not directly reflect library complexity. However, all three kits showed similar target coverage and uniformity of target coverage.
Table 6. Data from targeted sequencing libraries created with xGen DNA Library Prep Kit MC followed by xGen Pan-Cancer Hyb Panel (upper) or xGen Exome Hyb Panel (lower) capture reactions.
| Prep | Capture run | Input (ng) | Pre-Hyb yield (ng/ul) | Total reads | Mean insert (bp) | Mean coverage | Duplicates (%) | Estimated library | Fold 80 base penalty* | 50X Coverage (%) | 100X Coverage (%) | On-Target (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| xGen DNA Library Prep MC | xGen Pan-Cancer Hyb Panel MiSeq® | 10 | 97.2 | 1.5 M | 201 | 205X | 9.4 | 10 X 106 | 0 | 99.1 | 99 | 80.3 |
| 110 | 201 | 206X | 8.8 | 11 X 106 | 99.1 | 98.9 | 80.3 | |||||
| Supplier A | 101 | 201 | 203X | 9.6 | 10 X 106 | 99.2 | 99 | 80.2 | ||||
| 96 | 201 | 200X | 10.6 | 9 X 106 | 99.2 | 99 | 80 | |||||
| Supplier B | 99.6 | 196 | 203X | 10.1 | 9 X 106 | 99.1 | 99 | 80.3 | ||||
| 105 | 196 | 203X | 9.9 | 9 X 106 | 99.1 | 98.9 | 80.2 | |||||
| XGen DNA Library Prep MC | xGen Exome Hyb Panel NovaSeq® | 100 | 100 | 80 M | 212 | 146X | 9.1 | N/A | 1.53 | 85.6 | 77 | 89.1 |
| 110 | 211 | 146X | 8.2 | 1.48 | 85.9 | 78 | 88.6 | |||||
| Supplier A | 96.5 | 211 | 143X | 9.1 | 1.58 | 85.6 | 76 | 88.7 | ||||
| 101 | 213 | 141X | 9 | 1.54 | 85.8 | 76 | 88.2 | |||||
| Supplier B | 96 | 201 | 145X | 8.2 | 1.57 | 85.6 | 76 | 89.1 | ||||
| 98.4 | 207 | 141X | 9 | 1.55 | 85.8 | 76 | 88.7 |
* Fold 80 Base Penalty is the fold over-coverage necessary to raise 80% of bases in targets to the mean coverage.
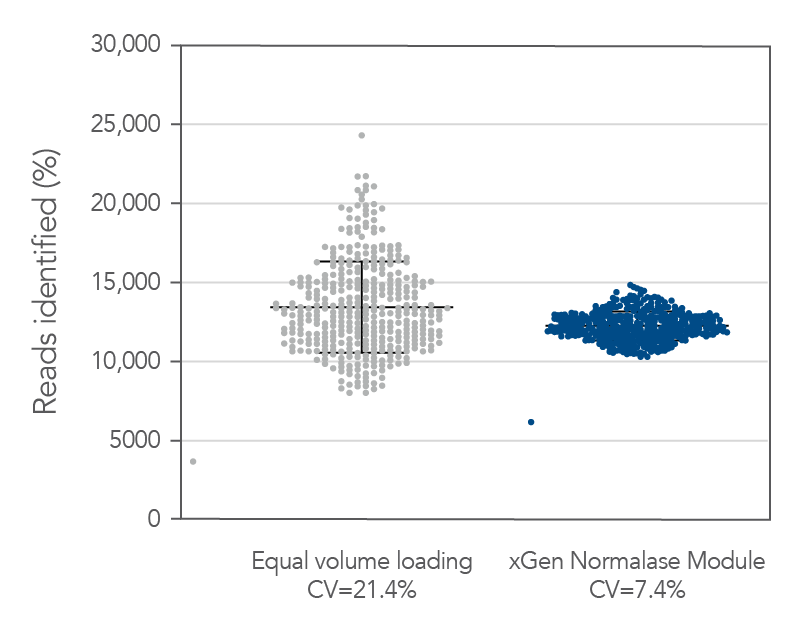
Figure 5. Normalase technology. 384 xGen DNA MC libraries were generated with 1 ng Coriell NA12878 gDNA and were each uniquely indexed with xGen Normalase Unique Dual Indexing primers during library amplification. Libraries were pooled with equal volumes, following PCR. The pool was quantified by Qubit™ (Thermo Fisher) and loaded on an Illumina MiSeq® system to obtain the percent reads Identified from each index. The equal volume pools coefficient of variation (CV) was 21%, showing amplification with the xGen Normalase indexing primers. The same libraries were then enzymatically normalized using xGen Normalase Module to generate an equimolar library pool, at a 4 nM concentration, and were then loaded on an Illumina MiSeq system. The Normalase pool CV as reduced to 7.4%, showing normalization of multiplexed libraries using xGen Normalase technology. Lines represent the median and 95% confidence interval.
Comprehensive metagenomics sequencing
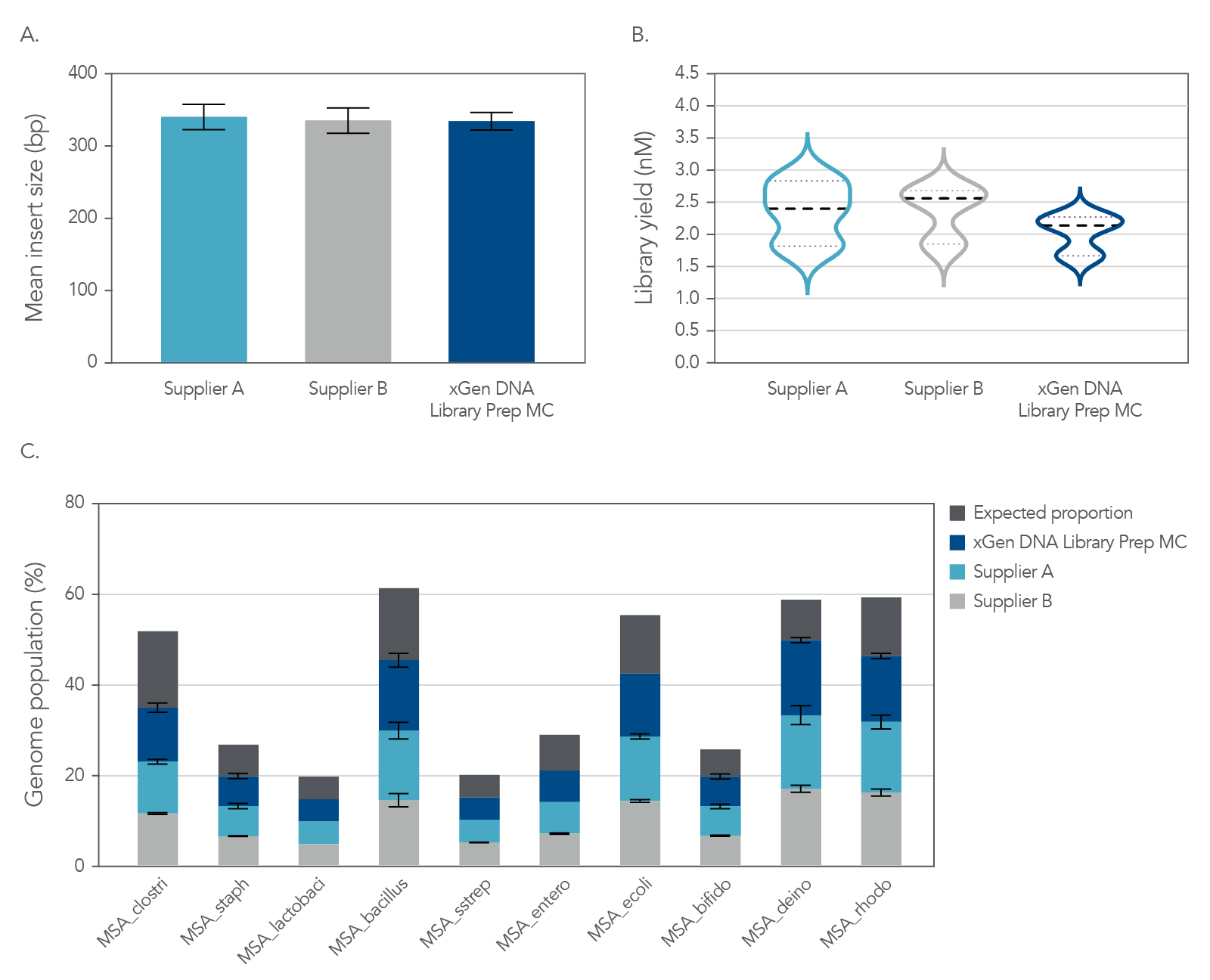
NGS libraries were constructed from 1 ng of mock metagenome DNA (ATCC MSA-1000) that was Covaris-sheared to 350 bp, in duplicate, using xGen DNA Library Prep MC UNI and two other suppliers’ kits. Libraries were prepared using IDT for Illumina TruSeq UDI DNA Indexes (Cat. No. 20020590) at supplier-specified adapter concentrations and were amplified with the polymerase supplied in each kit. Libraries were quantified by Qubit™ (Thermo Fisher) and Agilent Bioanalyzer™ 2100 and sequenced on an Illumina MiSeq system with 150 bp, paired end reads. Data (Figure 7) for each library were normalized to 1 M reads and achieved a metagenome mean coverage of 20X.
Figure 6. Comprehensive metagenomics sequencing. Top panel: Size selecting libraries using the same SPRI ratio resulted in a balanced comparison of the sequencing results. Comparable mean insert sizes and duplication rates were obtained with xGen DNA Library Prep MC UNI and two kits from other suppliers. Bottom panel: Despite significant variation in base composition, the xGen DNA Library Prep MC UNI and the other suppliers’ kits enabled identification of each strain’s genome at the expected frequency, with minimal bias. Variability in GC composition and genome size in this experiment supported that it did not influence results from the xGen™ DNA Library Prep MC UNI kit. The mock bacterial community contained the following strains: B. cereus (GC% = 35.5), B. adolescentis (GC% = 59.4), C. beijerinckii (GC% = 29.9), D. radiodurans (GC% = 66.7), E. faecalis (GC% = 37.8), E. coli (GC% = 50.8), L. gasseri (GC% = 35.3), R. sphaeroides (GC% = 68.9), S. epidermidis (GC% = 32.0), S. mutants (GC% = 36.8).
Senior Genomics Technologist
Queensland University of Technology
After prepping 50 samples with a DNA kit from another vendor and getting poor-quality libraries, I tried IDT's xGen EZ DNA prep kit on the exact same samples and got perfect libraries. We have also switched to this kit for plant samples that have failed with other vendors kits and everyone has been happy with the data. I have had similar experiences with methylation libraries that worked better with IDT and RNA. Overall, the kits are easy to use and give great results and we have switched several of our preps over to them.
Resources
Frequently asked questions
What are the DNA input requirements for the xGen DNA Library Prep Kits?
For PCR-free library construction, a minimum input of 100 ng of dsDNA is required for all xGen DNA Library Prep Kits.
For PCR-amplified library construction, xGen DNA Library Prep EZ and EZ UNI Kits supports dsDNA input range of 100 pg to 1 µg; while the xGen DNA Library Prep MC and MC UNI Kits support a dsDNA input range of 1 ng to µg.
What are the new names of IDT next generation sequencing products?
After IDT acquired Swift Biosciences, some of the IDT next generation sequencing products were renamed.
The following table lists the previous IDT product name corresponding with its new name, hyperlinked to the appropriate product page.
| Previous IDT product name | New IDT product name |
|---|---|
| xGen™ Prism™ DNA Library Prep Kit | xGen cfDNA & FFPE DNA Library Prep MC |
| COVID-19 Capture Bundle | xGen SARS-CoV-2 Hyb Panel Bundle |
| xGen CNV Backbone Panel—Tech Access | xGen CNV Backbone Hyb Panel |
| xGen Human ID Research Panel | xGen Human ID Hyb Panel |
| xGen Human mtDNA Research Panel v1.0 | xGen Human mtDNA Hyb Panel |
| xGen Exome Research Panel v2 | xGen Exome Hyb Panel v2 |
| Lotus™ DNA Library Prep Kit | xGen DNA Library Prep Kit MC or xGen DNA Library Prep Kit EZ |
| Custom or predesigned rhAmpSeq™ Library Prep Kits | Discontinued, but technology is available via rhAmpSeq for CRISPR Analysis System |
| Discovery Pools | xGen Custom Hyb Panels and xGen Custom Hyb Panels—Accel |
| Lockdown™ Probes Functional QC Discovery Pool | xGen Custom Hyb Panels—Production (available with and without functional testing) |
| xGen Predesigned Gene Capture Pools | xGen Custom Hyb Panels |
What are the new names of the Swift products?
The Swift products were rebranded and now belong to the xGen™ NGS product line. The following lists the old Swift product name, and then the new name hyperlinked to its current product page.
| Swift product name | New IDT product name |
|---|---|
| Accel-NGS® Adaptase® Module for Single-Cell Methyl-Seq | xGen Adaptase™ Module |
| Normalase® Amplicon Panels (SNAP) Core | xGen Amplicon Core |
| SARS-CoV-2 Additional Genome Coverage Panel | xGen SARS-CoV-2 Expanded Amplicon Panel |
| SARS-CoV-2 S Gene Panel | xGen SARS-CoV-2 SGene Amplicon Panel |
| SNAP Set 1A Combinatorial Dual Indexing Primers | xGen Amplicon CDI Primers |
| Swift Combinatorial Dual Indexing Primers | xGen CDI Primers |
| Swift Normalase® Combinatorial Dual Indexing Primers | xGen Normalase CDI Primers |
| Accel-NGS 1S Plus DNA Library | xGen ssDNA & Low-input DNA Library Prep |
| Swift 2S Sonic DNA Library | xGen DNA Library Prep MC |
| Swift 2S Sonic Flexible DNA Library | xGen DNA Library Prep MC UNI |
| Swift 2S Turbo v2 | xGen DNA Library Prep EZ |
| Swift 2S Turbo Flexible v2 DNA Library | xGen DNA Library Prep EZ UNI |
| Accel-NGS Methyl-Seq DNA Library | xGen Methyl-Seq Library Prep |
| Swift Normalase | xGen Normalase Module |
| Swift Rapid RNA Library | xGen RNA Library Prep Kit |
| Swift RNA Library | xGen Broad-Range RNA Library Prep |
What is the new name of the xGen™ Lotus™ DNA Library Prep Kit?
IDT has expanded its standard DNA NGS library prep kit offerings to include:
- xGen DNA Library Prep Kit MC compatible with mechanical shearing
- xGen DNA Library Prep Kit EZ that includes the reagents for enzymatic fragmentation
Another alternative is the:
- xGen ssDNA & Low-Input DNA Library Prep Kit for degraded DNA samples (formerly known as Accel-NGS® 1S Plus DNA Library)
Is titration of adapters required for different input amounts with the xGen™ DNA Library Prep Kit EZ / MC?
xGen DNA Library Prep Kit EZ
For direct sequencing of libraries prepared using the xGen DNA Library Prep Kit EZ from DNA inputs <25 ng, adapter dilution is necessary to achieve low levels of adapter dimers.
| DNA input (ng) | Reagent W4 |
|---|---|
| ≥25 | No dilution |
| 10 | 10-fold (1:10) |
| 1 | 20-fold (1:20) |
xGen DNA Library Prep Kit MC
For direct sequencing of libraries prepared using the xGen DNA Library Prep Kit MC from DNA inputs <50 ng, adapter dilution is necessary to achieve low levels of adapter dimers. Adapter titration recommendations are outlined in the table below. Optimization may be required for diluting adapters of intermediate input scale.
| DNA input (ng) | Adapter |
|---|---|
| ≥50 | No dilution |
| 10 | 10-fold (1:10) |
| 1 | 20-fold (1:20) |
How many cycles of library amplification should be used for the xGen™ DNA Library Prep Kits?
Below are the recommended minimum PCR cycles for direct sequencing of libraries prepared from high-quality genomic DNA for each xGen DNA Library Prep Kits.
When using indexing primer kits, a minimum number of 3 PCR cycles is required to complete adapter sequences, irrespective of whether enough library is available after ligation.
For samples of compromised quality, additional cycles may be required.
xGen DNA Library Prep Kit EZ
| Input material (ng) | Minimum PCR cycles |
|---|---|
| ≥100 | 3 |
| 25 | 5 |
| 10 | 7 |
| 1 | 10 |
xGen DNA Library Prep Kit MC
| Number of cycles required to generate | ||
|---|---|---|
| Input material (ng) |
100 ng library | 1 µg library |
| 1000 | 0* | 2 |
| 500 | 0* | 2–4 |
| 250 | 0–2* | 4–6 |
| 100 | 2–3 | 6–7 |
| 50 | 3–5 | 7–8 |
| 25 | 5–7 | 8–10 |
| 10 | 7–9 | 11–13 |
| 5 | 9–11 | 13–14 |
| 2.5 | 11–13 | 14–16 |
| 1 | 13–15 | 17–19 |
Are there any safe stopping points throughout the xGen™ DNA Library Prep Kit MC workflow?
If necessary, the protocol may be safely paused after completion of the post-ligation clean-up.
Purified adapter-ligated library may be stored at –20 ºC before amplification, target capture and/or sequencing library amplification products may be stored in a similar way, but the post-amplification clean-up should be performed as soon as possible.
Which adapters can be used with the xGen DNA Library Prep EZ and MC Kits?
The xGen Stubby Y Adapter is provided as part of the xGen DNA Library Prep EZ and MC Kits. With the Stubby Y Adapter, Tru-Seq-compatible indexing primers are used to incorporate indexes by PCR.
Visit IDT's xGen NGS Adapters & Indexing Primers page to learn more about IDT adapters.
While Tru-Seq-compatible full-length Y adapters can be used with the xGen DNA Library Prep EZ and MC Kits to index libraries by ligation, the "UNI" specific kits are better suited because the P5/P7 library amplification primers are included.
What is the shelf life and recommended storage conditions for xGen™ DNA Library Preparation Kits?
The recommended shelf life of the xGen DNA Library Prep Kit EZ and xGen DNA Library Prep Kit MC is at least 6 months from the time the product is delivered when stored at −20ºC and handled according to the protocol. The enzymes provided in these kits are temperature sensitive so appropriate care should be taken during handling and storage.
Upon receipt, immediately store the kit at −20°C.
What can cause under-fragmentation of DNA?
Under digestion is caused by either improper fragmentation time—see the CoA or product box for recommended lot-specific fragmentation time for the supported insert sizes; incomplete mixing of the master mix before or when combined with the sample; or when there is additional EDTA in your DNA sample storage buffer.
For ideal fragmentation, make sure your DNA is stored or eluted in a buffered solution containing Low EDTA TE (0.1 mM), as provided in the xGen DNA Library Prep Kit EZ, to maximize fragmentation efficiency.
If your DNA is in a 1 mM EDTA TE elution buffer used in the final steps of the DNA extraction or purification process, a buffer exchange using a column or bead-based purification step is required.
To avoid a purification step, you can alternatively adjust the amount of Reagent K2 used in the Enzymatic Prep step to no more than 3x to attain the desired fragment length.
What are the major steps in the xGen™ DNA Library Prep Kit EZ library construction?
Enzymatic prep. Consists of fragmentation of dsDNA followed by end-repair and dA-tailing of the fragments—all performed in a single reaction.
Adapter Ligation. Stubby or full-length Y adapters are ligated to 3’-dA-tailed molecules.
Library amplification. Employs a high-fidelity, low-bias polymerase. For indexing using the ligation workflow, amplification primers can be used, but PCR is optional when using the minimum of 100 ng input. For indexing by PCR, stubby Y adapters are used during ligation and amplified with indexing primers to complete the adapter sequence and add sample indexes.
Is the xGen DNA Library Prep Kit MC automation friendly?
Library construction using the xGen DNA Library Prep Kit MC is designed to be readily automatable.
A 10% overage volume of reagents is supplied in both the 16 and 96 reaction kits to accommodate automation. IDT does not supply automated liquid handling instruments or consumables but collaborates with automation solution providers and customers to develop and qualify automated scripts for use of our kits, with liquid handling platforms routinely used in NGS library preparation.
Contact your instrument vendor or Contact us if you plan to use this protocol with your automated liquid handling system.
What are the common applications for the xGen™ DNA Library Prep MC Kit?
The xGen DNA Library Prep MC Kit can be applied, but not limited to, the following applications:
- Whole genome sequencing (WGS)
- Targeted sequencing by hybridization capture (exome)
- Metagenomic sequencing
- Identification of germline inherited SNVs and Indels
- Identification of low-frequency somatic SNVs and Indels
- Copy number variation identification