Cas12a proteins
Harness the power of the Alt-R CRISPR-Cas12a enzymes to target genomic regions beyond the reach of Cas9. Explore our range of Cas12a (Cpf1) nucleases, including the proprietary Alt-R Cas12a Ultra, designed for enhanced editing precision, especially in AT-rich genomes. Achieve effective editing with staggered 5' overhangs and streamlined crRNA complexing—no tracrRNA required. Available for L.b. and A.s. Cas12a crRNAs.
Ordering
- Access additional genomic sites not available with Cas9, ideal for editing AT-rich genomes.
- Proprietary Alt-R Cas12a Ultra nucleases offering enhanced editing efficiency and precision, with activity at low temperatures—perfect for editing in ectothermic organisms.
- Reduce off-target editing with Alt-R Cas12a Ultra nucleases, making them ideal for precise and sensitive genome editing applications
Custom CRISPR solutions
Don’t see what you’re looking for? We are continually expanding our CRISPR product line, and we may have what you need. If you are interested in custom libraries, other CRISPR enzymes, formulations, or other CRISPR tools, contact our CRISPR experts today to discuss customized solutions for your research: CRISPR@idtdna.com.
Product Details
Cas12a (Cpf1) proteins
CRISPR-Cas12a (Cpf1) is an RNA-guided DNA endonuclease that is an alternative to the commonly used Streptococcus pyogenes Cas9 (S.p. Cas9) enzyme. Unlike S.p. Cas9, which recognizes NGG PAM sequences, Cas12a recognizes TTTV (V = A/G/C) PAM sites, thereby permitting genome editing in organisms with AT-rich genomes. A.s. Cas12a is an attractive option for genome editing applications due to its AT-rich PAM sequence, its highly specific DNA recognition and cleavage mechanism, and its native reliance on a single, short guide RNA.
Alt-R A.s. Cas12a (Cpf1) V3 nuclease
Alt-R A.s. Cas12a (Cpf1) Nuclease V3 enzyme is a high purity, recombinant Acidaminococcus sp. BV3L6 Cas12a. It is useful for targeting AT-rich regions when the Cas9-specific PAM sequence (NGG) is not available. The enzymes include nuclear localization sequences (NLSs) and C-terminal 6-His tags. The Cas12a enzyme must be combined with a gRNA to produce a functional, target-specific editing complex. For the best editing, combine Alt-R A.s. Cas12a (Cpf1) Nuclease V3 enzyme with optimized Alt-R CRISPR-Cas12a (Cpf1) crRNA in equimolar amounts.
Attention: Unlike S. pyogenes Cas9, which cleaves most NGG PAM sites to some degree, some of the tested TTTV sites show no cleavage by A.s. Cas12a nuclease. We recommend using positive control crRNAs to establish that your cells can be edited by Cas12a. In addition, we suggest testing 3 or more crRNAs per target gene.
Alt-R A.s. or L.b. Cas12a (Cpf1) Ultra Nucleases
The Alt-R Cas12a (Cpf1) Ultra Nucleases are also useful for targeting AT-rich regions without available Cas9-specific PAM sequences. However, they have much higher on-target potency than wild-type A.s. Cas12a (Cpf1). The Alt-R Cas12a (Cpf1) Ultra also can recognize many TTTT PAM sites in addition to TTTV motifs, increasing target range for genome editing studies. Furthermore, the new Alt-R Cas12a (Cpf1) Ultra nucleases are active at room temperature, making them flexible tools for applications requiring delivery at lower temperatures.
Comparison of CRISPR genome editing using Cas9 vs. Cas12a (Cpf1)
| Cas9 system | Cas12a system | |
|---|---|---|
| Applications |
|
|
| Ribonucleoprotein components |
|
|
| Variants |
|
|
| Cas9 crRNA:tracrRNA (option 1) | crRNA
tracrRNA
| — |
| Cas9 sgRNA (option 2) |
| — |
| Cas12a crRNA | — |
|
| CRISPR enzyme |
|
|
| DNA cleavage |
|
|
| PAM sequence† |
|
|
| Current recommendations for Alt-R RNP delivery |
|
|
* Molecular weight of Alt-R nuclease
† N = any base; V = A, C, or G
Product Data
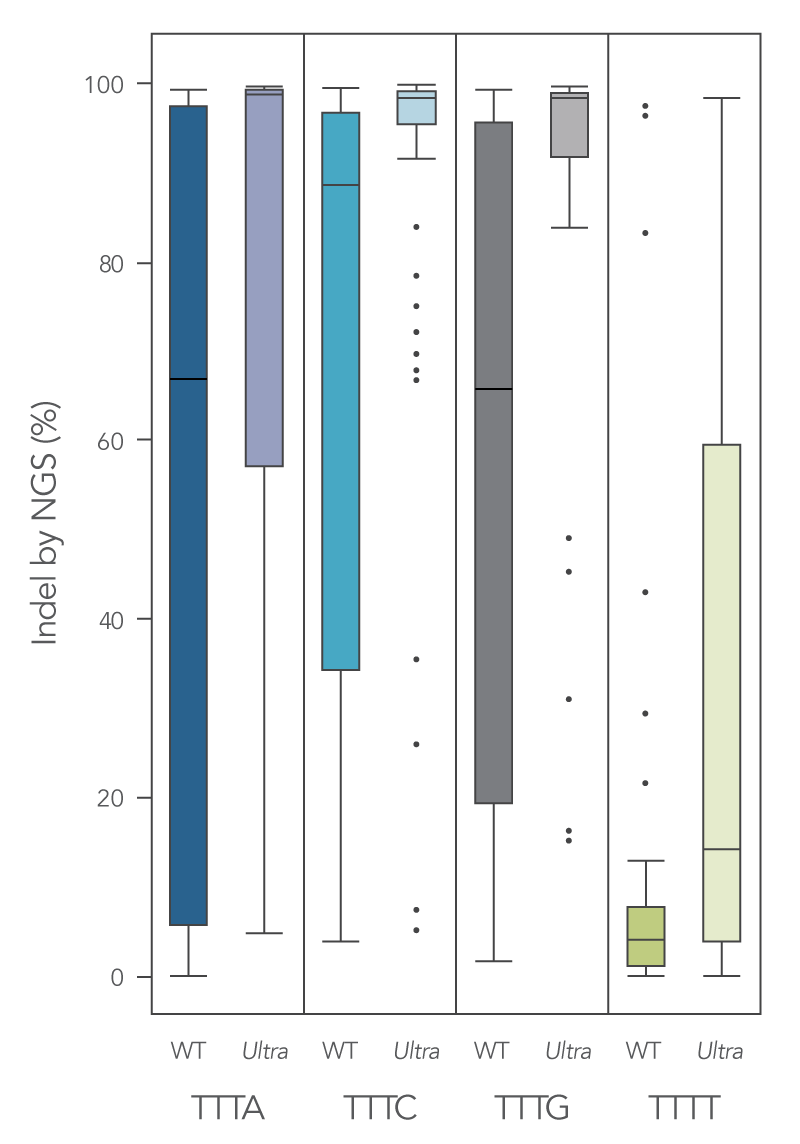
Newly developed Alt-R Cas12a (Cpf1) Ultra enzyme increases overall editing efficiency
To enhance activity, we introduced multiple modifications to the Cas12a protein that support notable improvement in overall editing efficiency. The new Alt-R Cas12a (Cpf1) Ultra nuclease has higher on-target potency than the wild-type A.s. Cas12a (Cpf1). The new Alt-R Cas12a (Cpf1) Ultra also can recognize many TTTT PAM sites in addition to TTTV motifs, increasing target range for genome editing studies (Figure 7). Furthermore, the new Alt-R Cas12a (Cpf1) Ultra nuclease is active at room temperature, making it a flexible tool for applications requiring delivery at lower temperatures.
Figure 1. New A.s. Cas12a (Cpf1) Ultra exhibits increased genomic editing efficiency in Jurkat and HEK-293 cells. Ribonucleoprotein (RNP) complexes were formed with wild type (WT) or Alt-R A.s. Cas12a (Cpf1) Ultra (Ultra), combined with crRNAs synthesized for 120 genomic loci to be delivered in Jurkat cells and 96 genomic loci to be delivered in HEK-293 cells. RNP complexes (4 μM) were delivered into Jurkat and HEK-293 cells via a Nucleofector™ system (Lonza) in the presence of Alt-R Cas12a (Cpf1) Electroporation Enhancer. Genome editing efficiencies were determined by target amplification followed by next generation sequencing on an Illumina instrument. The Cas12a-associated PAM sequences are indicated below the graph. n = 426, with 213 data points for WT and 213 data points for Cas12a Ultra. Each dot represents a single sample.
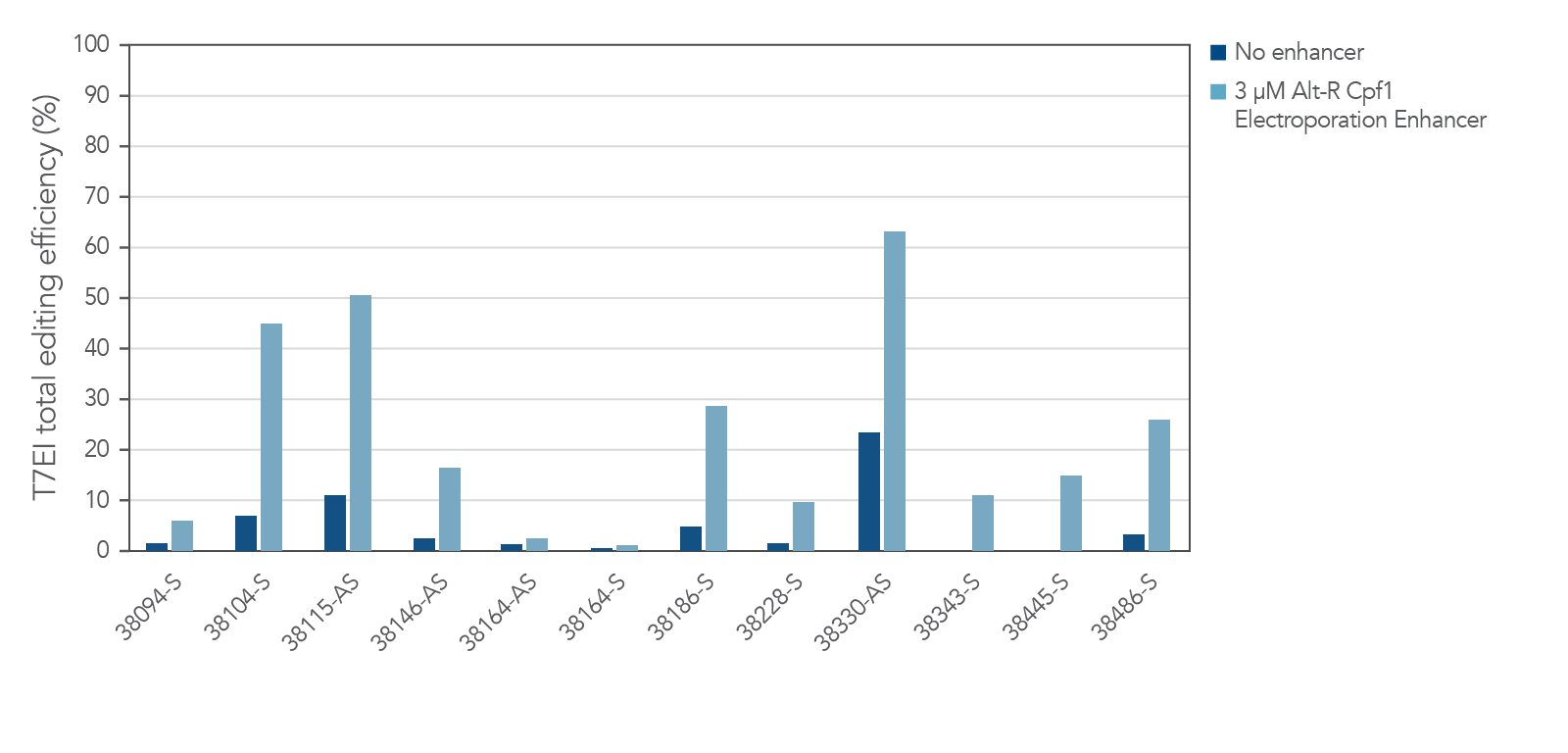
The electroporation enhancer is recommended for efficient genome editing with the CRISPR-Cas12a (Cpf1) system
Figure 2. Alt-R Cas12a (Cpf1) Electroporation Enhancer is required for efficient CRISPR editing in ribonucleoprotein (RNP) electroporation experiments. HEK-293 cells were electroporated with 5 µM RNP (Alt-R A.s. Cpf1 Nuclease 2 NLS complexed with Alt-R CRISPR-Cas12a (Cpf1) crRNA) as instructed in the Alt-R CRISPR-Cas12a (Cpf1) User Guide—RNP electroporation, Nucleofector™ system (available at www.idtdna.com/CRISPR-Cpf1). Twelve Cas12a PAM sites in the HPRT gene were targeted by Alt-R CRISPR-Cas12a (Cpf1) crRNAs. The electroporation reactions contained either no (dark blue) or 3 µM (light blue) Alt-R Cas12a (Cpf1) Electroporation Enhancer. Editing efficiency (n =3) was determined 48 hr after electroporation using the Alt-R Genome Editing Detection Kit, which provides the major components required for T7EI endonuclease assays. PAM = protospacer adjacent motif (Cas12a PAM sequence is TTTV); x-axis: numbers specify gene locations; S = sense strand; AS = antisense strand.
Resources
Frequently asked questions
What synthetic arrayed library formats are available from IDT? Are additional customization options possible?
- Table 1 shows available customizations for library design, formulations, and shipping formats.
Product Specifications
| Features | Options |
|---|---|
| Design | Predesigned, custom, user-provided |
| CRISPR systems | Cas9, Cas12a, Cas13, prime editing, and other alternative systems |
| Guaranteed Yield | 0.5 nmol, 2 nmol, 5 nmol, and custom normalized deliverables |
| Cas9 gRNA formats | 2-part cRNA:tracrRNA complex and sgRNA |
| Custom lengths supported | 30-150 nt |
| Chemical modifications | 2'-O-methyl RNA, PS linkages, end-blocking Alt-R modifications |
| Plate types | 96- & 384-well PCR, Deep-well, V-bottom, ECHO, custom options available |
| Formulation options | Multi-guide per well; pooled by gene Arrayed (single gRNA/well) Custom formulations upon request |
| QC | Individual ESI/MS |
| Supporting reagents & functional analysis pipeline (optional) | WT Cas9, HiFi Cas9, Cas12a, and Cas12a Ultra Glycerol-free options available in tubes or plates (ideal for robotics) Electroporation Enhancers rhAmpSeq™ CRISPR Analysis System (NGS-based on-/off-target editing analysis) |
- Don’t see what you’re looking for? We are continually expanding our CRISPR library products and we may have what you need. If you are interested in other chemically modified gRNAs (such as CRISPR on/off systems) targeting any sequence from any species, email our CRISPR experts today to discuss customized solutions for your research at CRISPR@idtdna.com.
What is the difference between A.s. and L.b. Cas12a? When should I use one or the other?
A.s. Cas12a is from Acidaminococcus sp. BV3L6, and L.b. Cas12a is from Lachnospiraceae bacterium. Both utilize the same TTTV PAM site and crRNA design considerations.
Both Cas12a nucleases result in robust editing at target sites at 37°C, with L.b. Cas12a also resulting in high editing efficiencies in plant genomes and at lower temperatures, such as 23°C.
What reagents will I need in addition to the Alt-R™ RNAs for a CRISPR-Cas12a (Cpf1) genome editing experiment?
The Alt-R™ CRISPR-Cas12a (Cpf1) System includes a target-specific CRISPR-Cas12a crRNA, Cas12a (Cpf1) endonucleases (Alt-R A.s.‑ Cas12a (Cpf1) V3, Alt-R A.s. Cas12a (Cpf1) Ultra, or Alt-R L.b. Cas12a (Cpf1) Ultra), and carrier DNA (Alt-R Cpf1 Electroporation Enhancer).
You will need to supply your own electroporation reagents to effectively deliver CRISPR reagents into your cell lines.
We recommend delivery of these reagents as a ribonucleoprotein (RNP). However, Alt-R A.s. Cas12a crRNA also works well in cells that stably express Acidaminococcus sp. BV3L6 Cpf1 nuclease.
We also highly recommend the use of control crRNAs. Control sequences for the CRISPR-Cas12a (Cpf1) system are provided online and in the user guides accessible here.
What methods can be used to deliver the Cas12a ribonucleoprotein (RNP)?
We have successfully used electroporation in internal experiments to deliver Cas12a RNP complexes to cells in culture. In many cell lines, Alt-R™ Cas12a Electroporation Enhancer, a non-targeting carrier DNA, is also required for efficient delivery of the RNP complex. IDT protocols using Cas12a and electroporation are included here, and these Cas12a DECODED articles include functional data and details:
What is the recommended length for the CRISPR-Cas12a crRNA guide?
For the Alt-R™ CRISPR-Cas12a crRNA, the input sequence can vary between 20 and 24 nucleotides. However, based on internal studies, we recommend using a 21-nucleotide sequence as input into the CRISPR-Cas12a crRNA ordering tool.
The ordering tool will automatically convert the DNA sequence to RNA during the ordering process. The 20- or 21-base constant region (loop domain) for the selected Cas12a nuclease (A.s. or L.b. Cas12a) and modifications that confer intracellular nuclease resistance will automatically be added. The final Alt-R CRISPR-Cas12a crRNA will be 40–45 bases.
What is the protospacer adjacent motif (PAM) sequence for Cas12a (Cpf1)? How is this different from Cas9?
The PAM sequence for the Cas12a (Cpf1) system is TTTV [1], where "V" is a A, C, or G. The Cas12a PAM sequence can be advantageous when working with T-rich target sequences. In contrast, the Cas9 PAM sequence is NGG.
Reference
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759-771.
What is Alt-R™ Cas12a (Cpf1) Electroporation Enhancer?
Alt-R Cas12a (Cpf1) Electroporation Enhancer improves electroporation efficiency and is a single-stranded DNA oligonucleotide that was computationally designed to be non-homologous to human, mouse, or rat genomes.
The use of this enhancer is important for efficient electroporation of ribonucleoproteins (RNPs), which, in turn, is important for increased rates of editing.
What buffer should I use to resuspend the Cas12a protein?
All Cas12a variants are provided as a 10 mg/mL solution and, therefore, resuspension is not required.
Is the Alt-R™ CRISPR-Cas12a ribonucleoprotein (RNP) complex stable? Do I need to make a fresh complex for each experiment?
Stability studies for the Alt-R Cas12a RNP complex have found that the Cas12a RNP complex is stable for up to 2 months at 4°C and for up to 1 year at –20°C.
Additional details are included in our CRISPR reagents stability DECODED article.
Why can't I see CRISPR-Cas12a (Cpf1) editing?
Check that you have included the Alt-R™ Cpf1 Electroporation Enhancer (carrier DNA) in the electroporation and that you are using one of our positive control crRNAs to monitor delivery efficiency.
Also check that the target site in your cells does not show any polymorphism that could affect the potency of the crRNA.
Target additional PAM sites in your gene of interest to identify a site that provides optimal editing efficiency.
How should I store the Cas12a protein?
Under optimal storage conditions, these proteins maintain functionality for at least 1-2 years. See our stability study for more details.
How should I deliver the CRISPR-Cas12a (Cpf1) components into cells?
Non-targeting carrier DNA (i.e., Alt-R™ Cpf1 Electroporation Enhancer) can also be included in the electroporation for efficient editing.
How much Alt-R™ Cas12a Electroporation Enhancer should I use?
We have observed that dose response curves vary for different guide sequences, depending on potency of the guide RNA. We recommend titrating the amount of ribonucleoprotein (RNP) and keeping the amount of electroporation enhancer fixed.
For the Nucleofector™ System (Lonza), we recommend 3 µM of Alt-R Cas12a Electroporation Enhancer, and for the Neon™ System (Thermo Fisher Scientific), we recommend 1.8 µM of Alt-R Cas12a Electroporation Enhancer. Toxicity may be observed at high concentrations of enhancer.
How do I design my CRISPR-Cas12a (Cpf1) crRNA guide sequence—correct orientation & strand?
Enter the 20–24 base DNA sequence downstream (3’ end) of the target Cas12a (Cpf1) PAM site (TTTV*), and then you are ready to continue. We typically recommend using a 21-nucleotide guide sequence.
The ordering tool will automatically convert the DNA sequence to RNA during the ordering process. If you are pasting your Cas12a (Cpf1) target site from an online design tool, make sure to verify the correct strand orientation before pasting the target sequence.
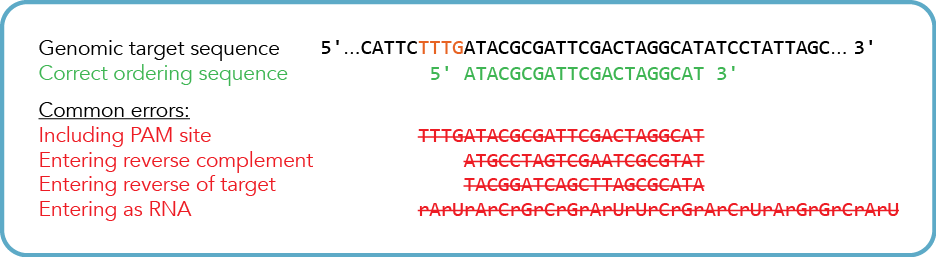
* V = A, C, or G