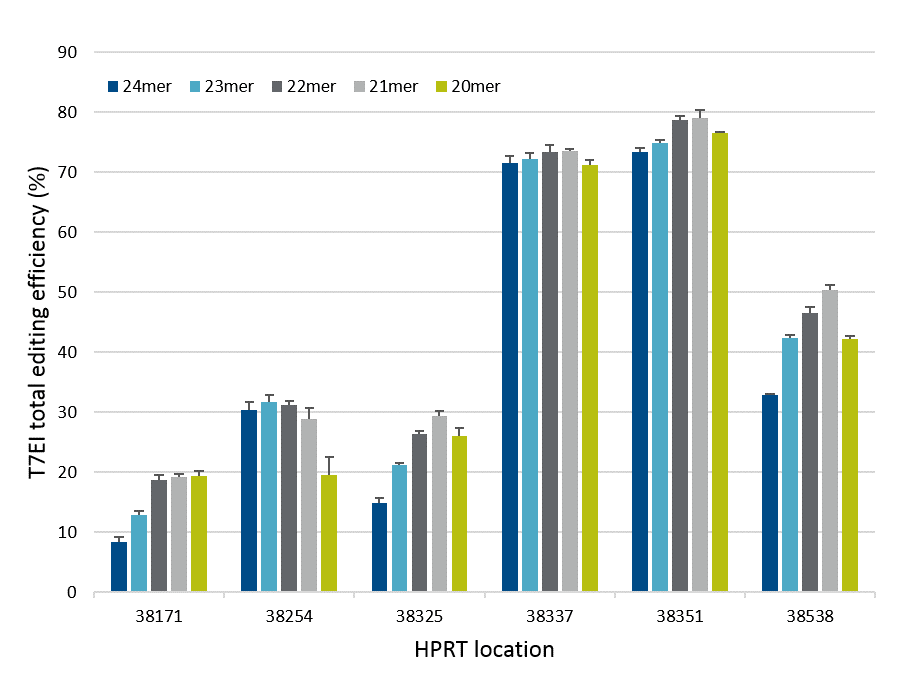Alt-R™ CRISPR-Cas12a (Cpf1) genome editing
Expand your research possibilities when genome editing.
The Alt-R CRISPR-Cas12a System enables targeting of additional genomic sites that are not available to the CRISPR-Cas9 System. We offer multiple Cas12a (Cpf1) enzymes, including IDT-proprietary Alt-R Cas12a Ultra nucleases with enhanced editing capabilities.
Ordering
- Enables genome editing in organisms with AT-rich genomes
- Allows interrogation of additional genomic regions compared to Cas9
- Produces staggered cut with 5’ overhang
- Simple complexing of crRNA with the Cas12a protein—no tracrRNA needed
- Support for L.b. and A.s. Cas12a crRNAs available in the ordering options
We recommend using Alt-R Cas12a (Cpf1) Ultra nucleases combined with Alt-R CRISPR-Cas12a (Cpf1) crRNA to generate a ribonucleoprotein (RNP) editing complex. The Alt-R Cpf1 Electroporation Enhancer is a Cas12a-specific carrier DNA reagent formulated for delivery by electroporation and is highly recommended for all experiments using this transfection method. View the Alt-R CRISPR-Cas12a (Cpf1) System protocols in the Resources below for guidance on electroporation and delivery of the RNP into your cell lines.
Customer-defined guide RNA
See Product details for design tips.
CRISPR-Cas12a crRNA
Customer-defined crRNA that will bind to 21–24 bases opposite to the TTTV, PAM sequence. Prices shown are for each crRNA. Plates require a minimum order of 24 crRNAs.
CRISPR-Cas12a crRNA in tubes
CRISPR-Cas12a crRNA in plates
CRISPR cGMP gRNA Manufacturing Service
Are you looking to accelerate your CRISPR genome editing program from discovery to clinical trials? Our Engineering Run and full cGMP guide RNA manufacturing services offer a streamlined and regulatory-compliant solution. Backed by comprehensive documentation, our guide RNAs simplify regulatory filings, offering a straightforward path to clinical success. Click here to learn more.
Custom CRISPR solutions
For more flexibility in guide RNA ordering, CRISPR Custom Guide RNAs allow a wide range of lengths and modifications to support a variety of CRISPR systems.
Don’t see what you’re looking for? We are continually expanding our CRISPR product line, and we may have what you need. If you are interested in custom libraries, other CRISPR enzymes, formulations, or other tools contact our CRISPR experts today to discuss customized solutions for your research: CRISPR@idtdna.com.
Cas12a Nuclease
The new Alt-R Cas12a (Cpf1) Ultra nucleases have higher on-target potency than their wild-type A.s. or L.b. Cas12a nuclease counterparts. These unique, proprietary mutants recognize many TTTT PAM sites in addition to TTTV motifs, which increases their target range for genome editing studies. In addition, our Alt-R L.b. Cas12a (Cpf1) Ultra mutant has been shown to have increased temperature tolerance, making it an ideal solution for systems that require lower culture temperatures, such as those needed for plant cultures.
The wild-type Alt-R A.s. Cas12a V3 enzyme is also available for legacy use.
Enhancers & control RNAs/primers
Control crRNAs for Cas12a (Cpf1)
Positive controls
To order, copy and paste the appropriate sequence in the CRISPR-Cas12a (Cpf1) crRNA Oligo Entry Tool:
Human HPRT1, Cas12a Positive Control crRNA: GGTTAAAGATGGTTAAATGAT
Mouse Hprt, Cas12a Positive Control crRNA: GGATGTTAAGAGTCCCTATCT
Rat Hprt1, Cas12a Positive Control crRNA: ATGCTTAAGAGGTATTTGTTA
Negative controls
To order, copy and paste the appropriate sequence in the CRISPR-Cas12a (Cpf1) crRNA Oligo Entry Tool:
Cas12a Negative Control crRNA #1: CGTTAATCGCGTATAATACGG
Cas12a Negative Control crRNA #2: CATATTGCGCGTATAGTCGCG
Cas12a Negative Control crRNA #3: GGCGCGTATAGTCGCGCGTAT
Product Details
Alt-R CRISPR-Cas12a System
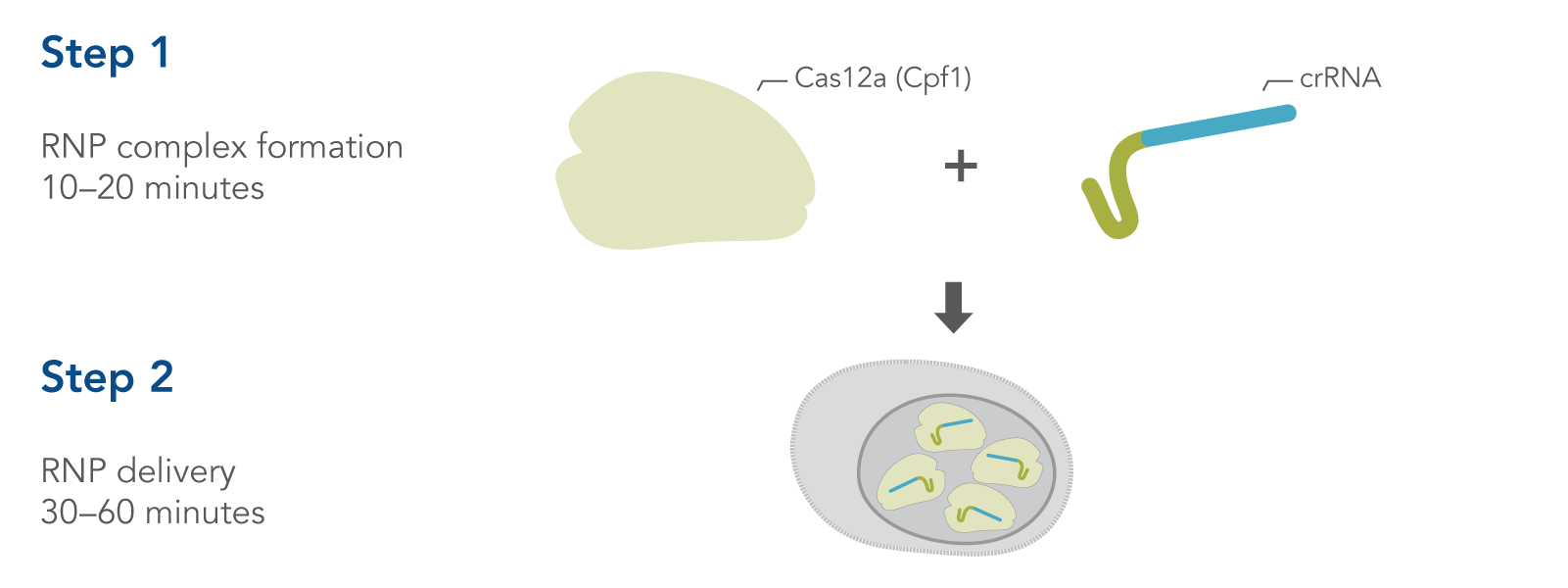
Simple, two-step delivery of ribonucleoprotein complexes (crRNA:Cas12a)
Figure 1. Overview of Alt-R CRISPR-Cas12a System experiments for ribonucleoprotein (RNP) delivery by electroporation.
CRISPR-Cas12a genome editing method uses the Cas12a endonuclease to generate double-stranded breaks that contain a staggered 5′ overhang. Cas12a requires only a single CRISPR RNA (crRNA) to specify the DNA target sequence (Figure 1). After cleavage, DNA is then repaired by non-homologous end-joining (NHEJ) or homology-directed recombination (HDR), resulting in a modified sequence. Alt-R CRISPR-Cas12a reagents provide essential, optimized tools needed to use this pathway for genome editing research. A brief comparison of CRISPR-Cas9 and CRISPR-Cas12a is provided at the end of this section.
| Alt-R CRISPR-Cas12a System | |
|---|---|
| Required components | |
| Alt-R CRISPR-Cas12a (Cpf1) crRNA | Target-specific RNA oligo, custom synthesized based on your sequence |
Alt-R A.s. or L.b. Cas12a (Cpf1) Ultra (recommended) |
|
| Alt-R Cas12a (Cpf1) Electroporation Enhancer | Cas12a-specific carrier DNA highly recommended for efficient electroporation of the RNP |
| Related reagents and kits | |
|---|---|
| Alt-R HDR Enhancer V2 | For improved rates of homology-directed repair |
| Cas12a positive controls | Order as custom crRNAs or oligos
|
| Alt-R Genome Editing Detection Kit | For mutation detection and estimating editing efficiency |
Components for genome editing
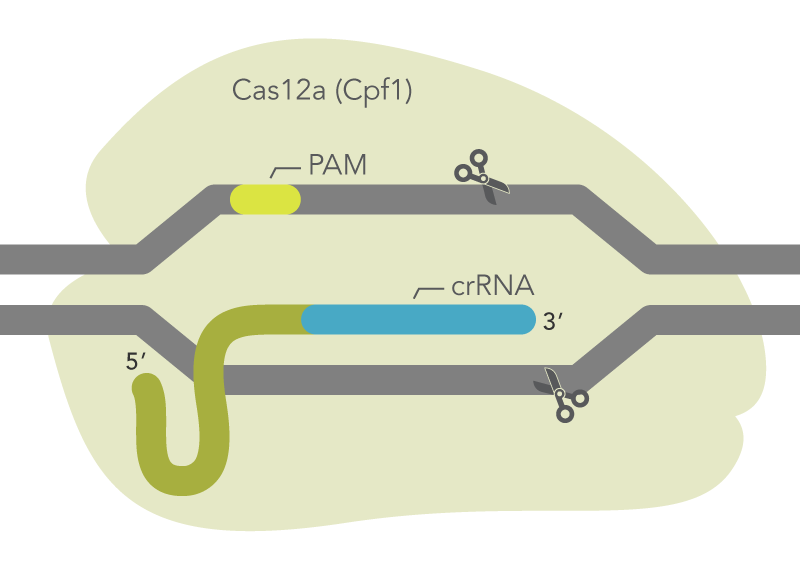
Cas12a endonucleases from Acidaminococcus sp. BV3L6 or Lachnospiracaea bacterium ND2006 along with a crRNA is capable of mediating genome editing in mammalian cells (Figure 2). The Alt-R CRISPR-Cas12a System includes three main components: an optimized crRNA, A.s. or L.b. Cas12a endonuclease, and Alt-R Cas12a (Cpf1) electroporation enhancer. While electroporation of Cas12a endonuclease as part of an RNP is the preferred method, the Alt-R CRISPR-Cas12a crRNA is also compatible with A.s. or L.b. Cas12a from any source, including cells that stably express A.s. Cas12a endonuclease.
Figure 2. Components of the Alt-R CRISPR-Cas12a System for directing Cas12a endonuclease to genomic targets. Alt-R Cas12a (Cpf1) crRNA forms a complex with Cas12a endonuclease to guide targeted cleavage of genomic DNA. The cleavage site is specified by the spacer element of the crRNA (blue bar). The crRNA spacer element recognizes 21 nt on the opposite strand of the TTTV PAM site. The PAM site must be present immediately upstream of the protospacer element for cleavage to occur. PAM = protospacer adjacent motif; V = A, C, or G
Alt-R CRISPR-Cas12a (Cpf1) crRNA and design tips
The Alt-R CRISPR-Cas12a (Cpf1) crRNA is a single, 40–44 base, guide RNA, comprising a 20 base constant region (loop domain) and a 20–24 base target-specific region (protospacer domain). We typically recommend a 21 base protospacer domain for optimal activity. All Alt-R CRISPR-Cas12a (Cpf1) crRNAs are synthesized with proprietary chemical modifications, which protect the crRNA from degradation by cellular RNases and further improve on-target editing performance. For crRNAs used with A.s. Cas12a, identify locations in your target region with the protospacer adjacent motif (PAM) sequence, TTTV, where V is A, C, or G. Your Alt-R CRISPR-Cas12a (Cpf1) crRNA will bind to the DNA strand opposite to the PAM sequence (Figure 2). Do not include the PAM sequence in your crRNA design. An example of a correct crRNA sequence is shown in Figure 3.
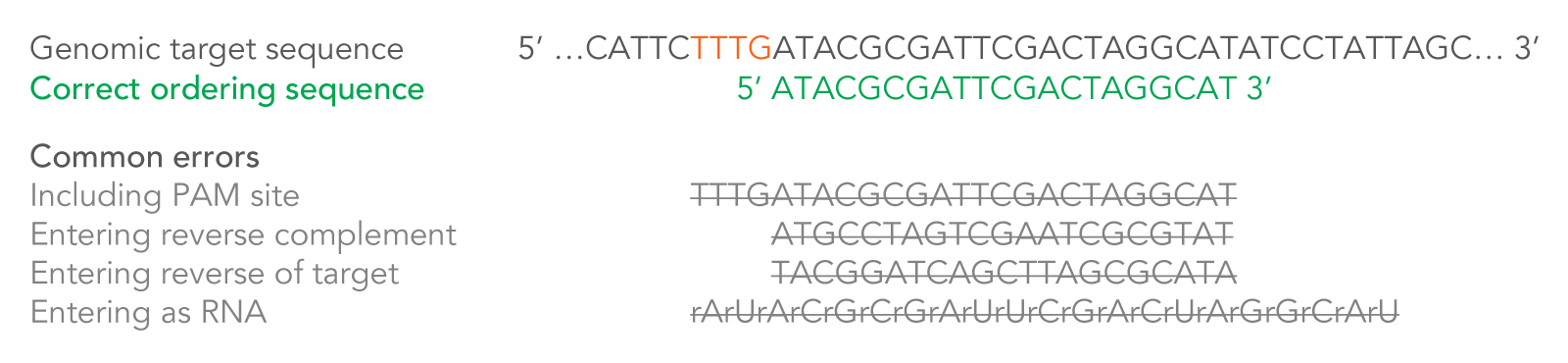
After you enter your 20–24 base target sequence, 20 additional bases and the necessary modifications will automatically be added by the order entry system for a total of 40–44 RNA bases. These additional bases and modifications are necessary to create a complete Alt-R CRISPR-Cas12a (Cpf1) crRNA. The system will also convert the final sequence to RNA—enter DNA bases only into the ordering tool (Figure 3).
Figure 3. How to enter your Cas12a (Cpf1) crRNA target sequence. Because the crRNA recognizes and binds 21 bases on the DNA strand opposite from the TTTV sequence of the PAM site, order your crRNA by entering the 20–24 bases downstream of the PAM site, in the forward orientation as shown. Enter only DNA bases into the order entry tool. If you are pasting your CRISPR target site from an online design tool, make sure you verify the correct strand orientation. Do not include the PAM site in your design. Common incorrect design examples are shown as crossed-out gray text. PAM = protospacer adjacent motif; V = A, C, or G
Alt-R A.s. Cas12a (Cpf1) nuclease
Alt-R A.s. Cas12a (Cpf1) Ultra and V3 nucleases are high purity, recombinant Acidaminococcus sp. Cas12a. The enzymes include nuclear localization sequences (NLSs) and C-terminal 6-His tags. The Cas12a enzyme should be combined with a crRNA to produce a functional, target-specific editing complex. For the best editing, combine Alt-R A.s. Cas12a Ultra or V3 enzyme with optimized Alt-R CRISPR-Cas12a crRNA in equimolar amounts.
In contrast to Streptococcus pyogenes Cas9, which recognizes an NGG PAM sequence, the A.s. Cas12a PAM sequence is TTTV, which permits targeting of DNA sequences in AT-rich regions of the genome. The Alt-R A.s. Cas12a Ultra enzyme has increased recognition of the TTTT PAM sequence.
Alt-R L.b. Cas12a (Cpf1) Ultra nuclease
Alt-R L.b. Cas12a (Cpf1) Ultra nuclease is high purity, recombinant Lachnospiraceae bacterium ND2006 (L.b.) nuclease, purified from an E. coli strain expressing Cas12a with proprietary mutations to improve both on-target performance and temperature tolerance. The enzyme includes nuclear localization sequences (NLSs) and C-terminal 6-His tags. The Cas12a enzyme should be combined with a crRNA to produce a functional, target-specific editing complex. For the best editing, combine Alt-R L.b. Cas12a Ultra enzyme with optimized Alt-R CRISPR-Cas12a crRNA in equimolar amounts.
L.b. Cas12a (Cpf1) Ultra mutant has been shown to have increased temperature tolerance for systems that require lower culture temperatures, such as those needed for plant cultures.
Alt-R Cas12a (Cpf1) Electroporation Enhancer
The Alt-R Cas12a (Cpf1) Electroporation Enhancer is a Cas12a-specific carrier DNA that is optimized to work with the Amaxa® Nucleofector® device (Lonza) and the Neon® Transfection System (Thermo Fisher) for increased transfection efficiency and therefore, increased genome editing efficiency. The electroporation enhancer is non-targeting and shows no integration into the target site based on next-generation sequencing experiments. The electroporation enhancer is highly recommended for optimal genome editing with the CRISPR-Cas12a system.
Alt-R™ HDR Enhancer Protein
Alt-R HDR Enhancer Protein is a novel protein-based reagent that promotes homology-directed repair by inhibiting 53BP1, a key regulator of dsDNA break repair pathway choice. It improves editing efficiency by up to 2X across established and hard-to-edit primary cells (iPSCs, HSPCs) without increasing off-target effects, genomic translocations, or compromising cell viability. Compatible with multiple CRISPR systems, including Cas9, Cas12a, and Eureca™-V, it integrates seamlessly into existing workflows. Offered in RUO format with a cGMP grade coming soon, Alt-R HDR Enhancer Protein is specifically engineered for translational researchers advancing CRISPR-based therapeutics.
Alt-R HDR Enhancer V2
Alt-R HDR Enhancer is a small molecule compound that increases homology-directed repair. Alt-R HDR Enhancer V2 is a new and improved small molecule compound that exhibits its activity in multiple cell lines, including both adherent and suspension cell lines. Its activity is independent of the enzyme employed; for example, it can be used either with Alt-R S.p. Cas9 nucleases or A.s. Cas12a (Cpf1) nucleases. This versatile reagent is also compatible with electroporation and lipofection methods.
Positive controls
crRNAs
Positive control crRNAs can be used to show that Cas12a editing is occurring in your experiments, which can be useful when you are finalizing ribonucleoprotein (RNP) delivery conditions or if you need to troubleshoot your experiments.
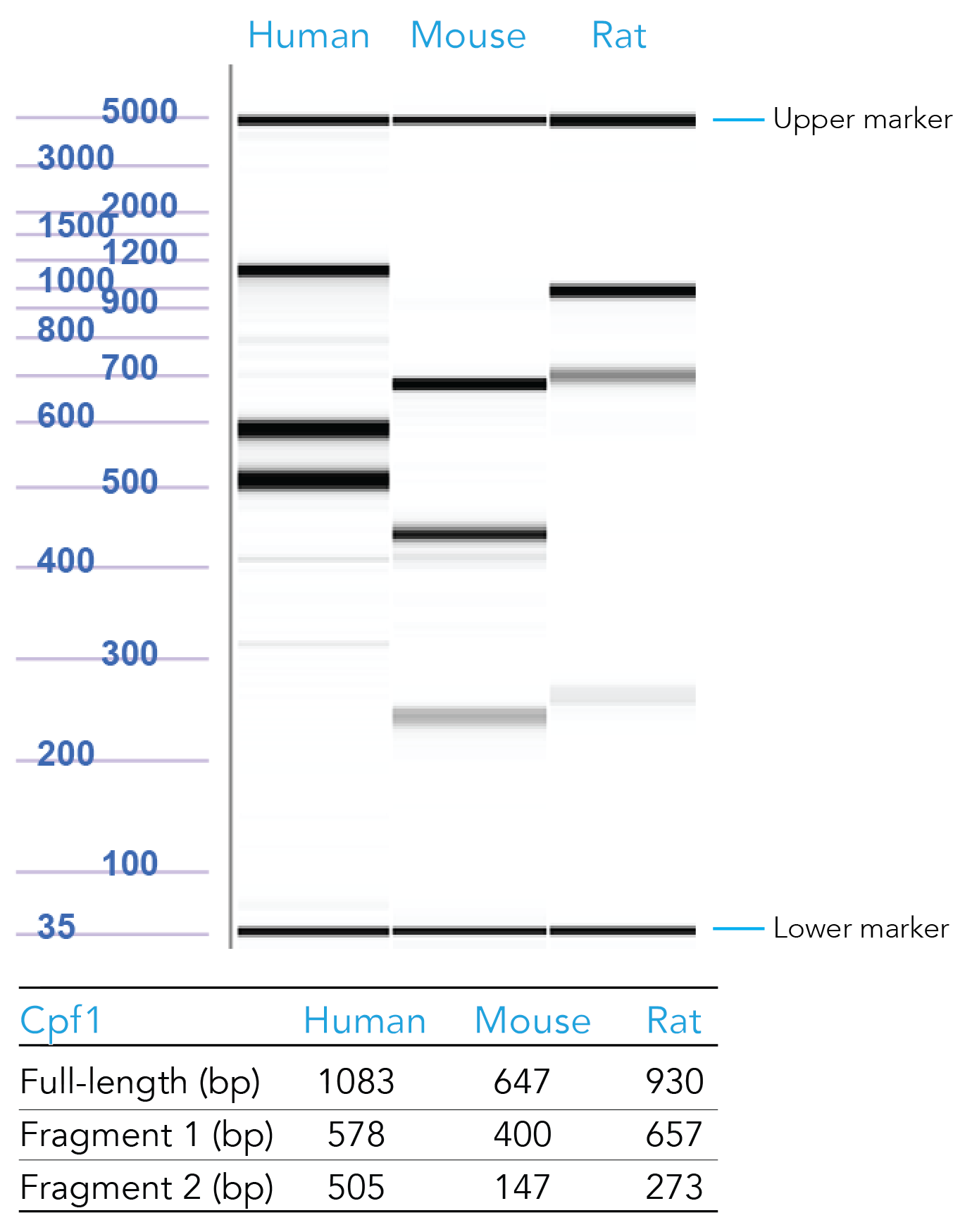
Our scientists have formulated positive control crRNAs targeting HPRT (Figure 4). To order, copy and paste the appropriate sequence in the Cpf1 crRNA ordering page:
Human HPRT1, Cas12a Positive Control crRNA: GGTTAAAGATGGTTAAATGAT
Mouse Hprt, Cas12a Positive Control crRNA: GGATGTTAAGAGTCCCTATCT
PCR primers
Our scientists have designed PCR primers (Alt-R HPRT PCR Primer Mixes for human or mouse) for use with the Alt-R Genome Editing Detection Kit to identify editing or estimate editing efficiency in samples transfected with the positive control HPRT crRNAs.
Figure 4. Sample data from T7 endonuclease I (T7EI) assays of samples transfected with Cas12a HPRT Positive Controls. Genomic DNA from CRISPR-Cas12a edited human, mouse, and rat HPRT controls were PCR amplified, digested using T7EI, and run on the Fragment Analyzer™ system (Advanced Analytical Technologies, Inc.). Reference standard bands at 5000 bp (upper marker) and 35 bp (lower marker) are used to align the gel and analyze the results. Estimated band sizes for the cut and uncut positive control amplicons are listed in the table. Cell lines used were HEK-293 (human), Hepa1-6 (mouse), and RG2 (rat). PCR annealing temperatures for human and mouse primers is 67°C and for rat primers is 64°C.
Negative controls
Negative control crRNAs are important for showing that transfection of the RNP complex is not responsible for observed phenotypes. Amplification of DNA from these negative control samples with your experimental primers and cycling conditions should result in only full-length products with the Alt-R Genome Editing Detection Kit (i.e., in a T7EI assay). Note that this result does not rule out off-target effects of your experimental crRNA.
Our scientists have computationally formulated negative control crRNAs to be non-targeting in human or mouse genomes. To order, copy and paste the appropriate sequence in the Cas12a (Cpf1) crRNA ordering page:
Cas12a Negative Control crRNA #1: CGTTAATCGCGTATAATACGG
Cas12a Negative Control crRNA #2: CATATTGCGCGTATAGTCGCG
Cas12a Negative Control crRNA #3: GGCGCGTATAGTCGCGCGTAT
Comparison of CRISPR genome editing using Cas9 vs. Cas12a (Cpf1)
| Cas9 system | Cas12a system | |
|---|---|---|
| Applications |
|
|
| Ribonucleoprotein components |
|
|
| Variants |
|
|
| Cas9 crRNA:tracrRNA (option 1) | crRNA
tracrRNA
| — |
| Cas9 sgRNA (option 2) |
| — |
| Cas12a crRNA | — |
|
| CRISPR enzyme |
|
|
| DNA cleavage |
|
|
| PAM sequence† |
|
|
| Current recommendations for Alt-R RNP delivery |
|
|
* Molecular weight of Alt-R nuclease
† N = any base; V = A, C, or G
Product Data
Alt-R Cas12a (Cpf1) Ultra enzymes plus supporting reagents increase overall editing efficiency
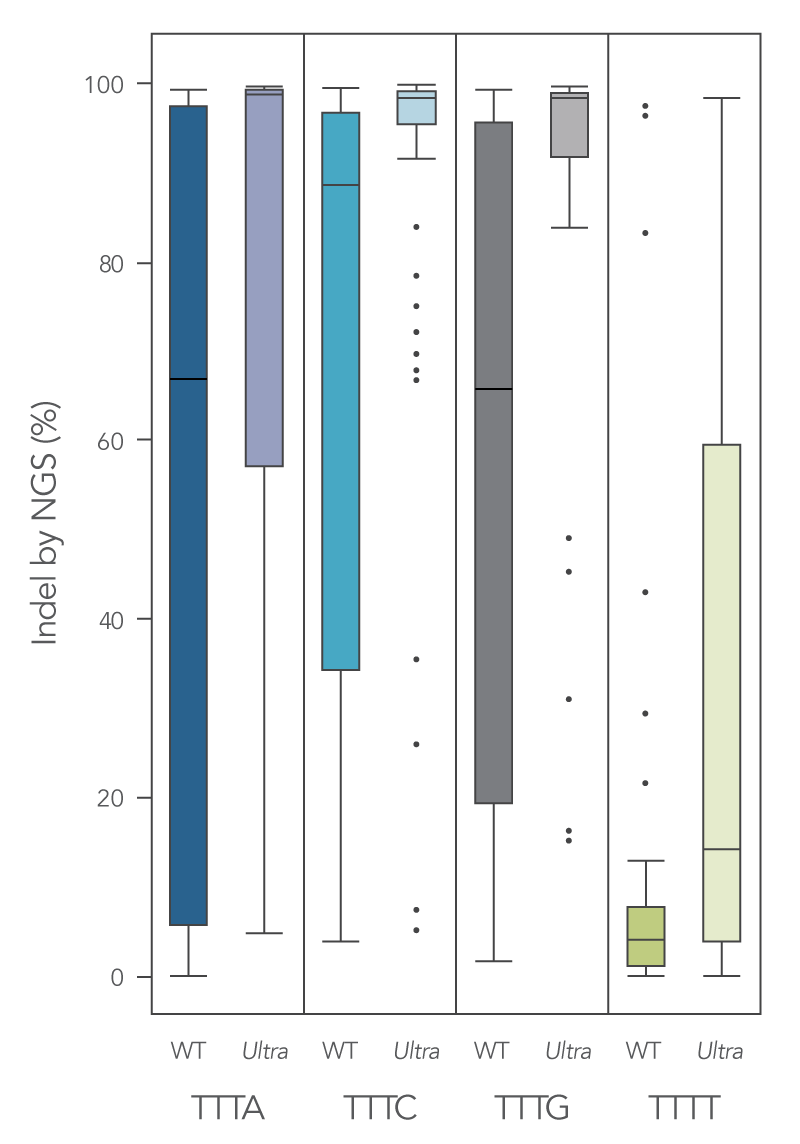
To enhance activity, we introduced multiple modifications to the Cas12a protein that resulted in measurable improvement in overall editing efficiency in comparison to the wild-type version. The new Alt-R A.s. Cas12a (Cpf1) Ultra nuclease has higher on-target potency than the wild-type A.s. Cas12a (Cpf1). Alt-R Cas12a (Cpf1) Ultra can also recognize many TTTT PAM sites in addition to TTTV motifs, increasing target range for genome editing studies (Figure 2). Furthermore, the Alt-R Cas12a (Cpf1) Ultra nuclease is active even at room temperature, making it a flexible tool for applications requiring delivery at lower temperatures.
Figure 1. New Alt-R A.s. Cas12a (Cpf1) Ultra exhibits increased genomic editing efficiency in Jurkat and HEK-293 cells. Ribonucleoprotein (RNP) complexes were formed with wild type (WT) or Alt-R A.s. Cas12a (CPF1) Ultra (Ultra), combined with crRNAs synthesized for 120 genomic loci to be delivered in Jurkat cells and 96 genomic loci to be delivered in HEK-293 cells. RNP complexes (4 μM) were delivered into Jurkat and HEK-293 cells via a Nucleofector™ system (Lonza) in the presence of Alt-R Cas12a (Cpf1) Electroporation Enhancer. Genome editing efficiencies were determined by target amplification followed by next-generation sequencing on an Illumina instrument. The Cas12a-associated PAM sequences are indicated below the graph.
Exact length of Cas12a (Cpf1) crRNA improves gene editing
Systematic variation of crRNA length led to the development of the Alt-R CRISPR-Cas12a (Cpf1) crRNA, which shows improved gene editing in mammalian cells. Overall, crRNAs containing 21
Figure 2. crRNA with 21mer protospacer lengths provides optimal on-target genome editing. Varying lengths of crRNAs targeting HPRT were reverse transfected, using Lipofectamine® RNAiMAX™ reagent (Thermo Fisher), into a HEK-293–Cas12a cell line that stably expresses Acidaminococcus sp. Cas12a. Genomic DNA was isolated, and genome editing was measured using the Alt-R Genome Editing Detection Kit (T7EI assay) and Fragment Analyzer (Advanced Analytical).
Resources
Frequently asked questions
What is the difference between A.s. and L.b. Cas12a? When should I use one or the other?
A.s. Cas12a is from Acidaminococcus sp. BV3L6, and L.b. Cas12a is from Lachnospiraceae bacterium. Both utilize the same TTTV PAM site and crRNA design considerations.
Both Cas12a nucleases result in robust editing at target sites at 37°C, with L.b. Cas12a also resulting in high editing efficiencies in plant genomes and at lower temperatures, such as 23°C.
What buffer should I use to resuspend the Cas12a protein?
All Cas12a variants are provided as a 10 mg/mL solution and, therefore, resuspension is not required.
How should I deliver the CRISPR-Cas12a (Cpf1) components into cells?
Non-targeting carrier DNA (i.e., Alt-R™ Cpf1 Electroporation Enhancer) can also be included in the electroporation for efficient editing.
Do you offer CRISPR gRNA libraries?
Yes, we offer predesigned Cas9 CRISPR crRNA and additional options for creating customizable gRNA libraries.
For more information, please visit our Custom guide RNA Libraries page.