Genetic disease research
Genetic disease research has greatly benefited from next generation sequencing, but challenges remain, such as needing to keep panels updated with recently discovered targets and coverage of variants important in diverse populations. Learn about these challenges and how Archer NGS assay solutions can help.
Archer™ NGS assay solutions
Overview
Genetic disease profiling challenges with NGS
Next generation sequencing (NGS) has enabled high-throughput screening of a multitude of variants useful for inherited disease research, enabling fast, sensitive identification of variants. In fact, NGS has been seen as a turning point in rare disease research, with the number of new rare disease-associated genes growing exponentially in recent years to over 4,900[1]. However, this rapid discovery also means it is difficult for labs to keep up with a relevant targeted regions in their NGS assays. Also, genome-wide association studies historically have focused mostly on variants common in European populations[2], this means assays may not be optimized for use in diverse populations. Therefore, it can be challenging for labs to know if they are missing germline variants in samples from diverse genetic ancestries.
Additionally, NGS workflows can be cumbersome and allow many points for manual errors and analysis tools can be expensive and time-consuming to maintain. Bioinformatic pipelines may also lack the QC and statistical tools necessary to guide calling and reduce errors, which can put the burden on lab staff to make difficult calls.
Genetic disease assays fit for your lab’s needs
Archer's NGS assays resolve these challenges with patented AMP chemistry, streamlined parallel workflows, and user-friendly Archer Analysis. Anchored Multiplex PCR (AMP™) is a target enrichment method that uses molecular barcoded adapters and single, nested, gene-specific primers for amplification, permitting open-ended capture of nucleic acids. This approach enables the detection of multiple variant types and protects against dropout of primers. Primers function independently, which means new gene targets can be easily added to existing panels without compromising assay performance. As an example, for the commonly-studied Cystic Fibrosis Conductance Regulator (CFTR) gene, the catalog VARIANTPlex CFTR v2 panel provides extensive coverage of variants relevant for diverse populations, including all coverage of exons and key intronic regions.
To fit your lab’s unique needs, Archer's genetic disease assays are available in two reagent formats:
- Liquid reagents are fit for high-throughput, liquid handling workflows. Liquid adapters are available in 96-well plate format and include an increased number of dual indexes (36,000 unique combinations of sequencing indices) to support high-throughput sequencing.
- Lyophilized reagents are fit for lower throughput, manual workflows. These single-use, reaction-size reagents allow your lab to avoid potential contamination during manual pipetting and provide extended shelf life.
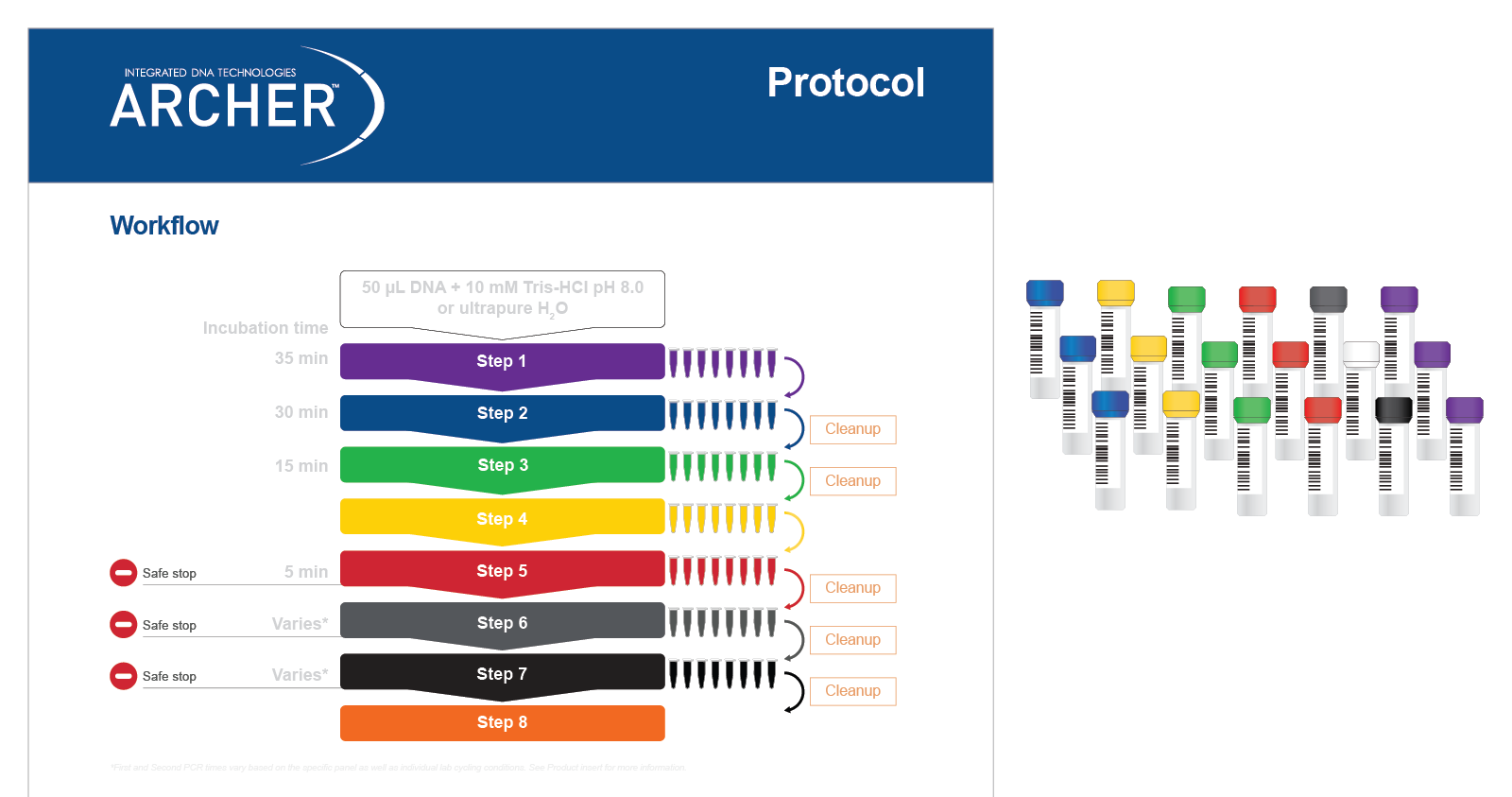
Regardless of reagent format, all reagents are color-coded to match protocol steps, making benchwork easy (Figure 1).
Genetic Disease Research Assays
Customize content for genetic disease panels
Customize the above assays by adding any of our functionally-tested designs or create a new panel that fits your exact requirements with Assay Marketplace.
Learn moreReady to start?
Talk with our technical sales team. Learn how our genetic disease research assays can identify key genetic alterations for your research.
Request a consultationReferences
- Fernández-Marmiesse A, Gouveia S, Couce ML. NGS Technologies as a Turning Point in Rare Disease Research, Diagnosis and Treatment. Curr Med Chem. 2018 Jan; 25(3): 404–432.
- Need AC, Goldstein AB. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25(11):489-94.


 Processing
Processing