CRISPR genome editing
The Alt-R CRISPR Systems were developed through comprehensive research on each component of the CRISPR-driven technology that generates a double-stranded break critical for gene disruption and DNA insertion by homologous recombination.
A complete workflow solution, from design to analysis.
Overview
Use of the CRISPR (clustered regularly interspaced short palindromic repeats) and associated Cas9 enzyme for genome editing has been a major technological breakthrough, making genome modification in cells or organisms faster, more efficient, and more robust than previous genome editing methods. The Alt-R CRISPR-Cas9 System is an optimized genome editing solution for producing on-target, double-stranded DNA breaks.
We have also developed an alternative Alt-R CRISPR-Cas12a (Cpf1) System to open up CRISPR editing to additional areas in genomes.
Quick comparison of CRISPR genome editing using Cas9 vs. Cas12a (Cpf1)
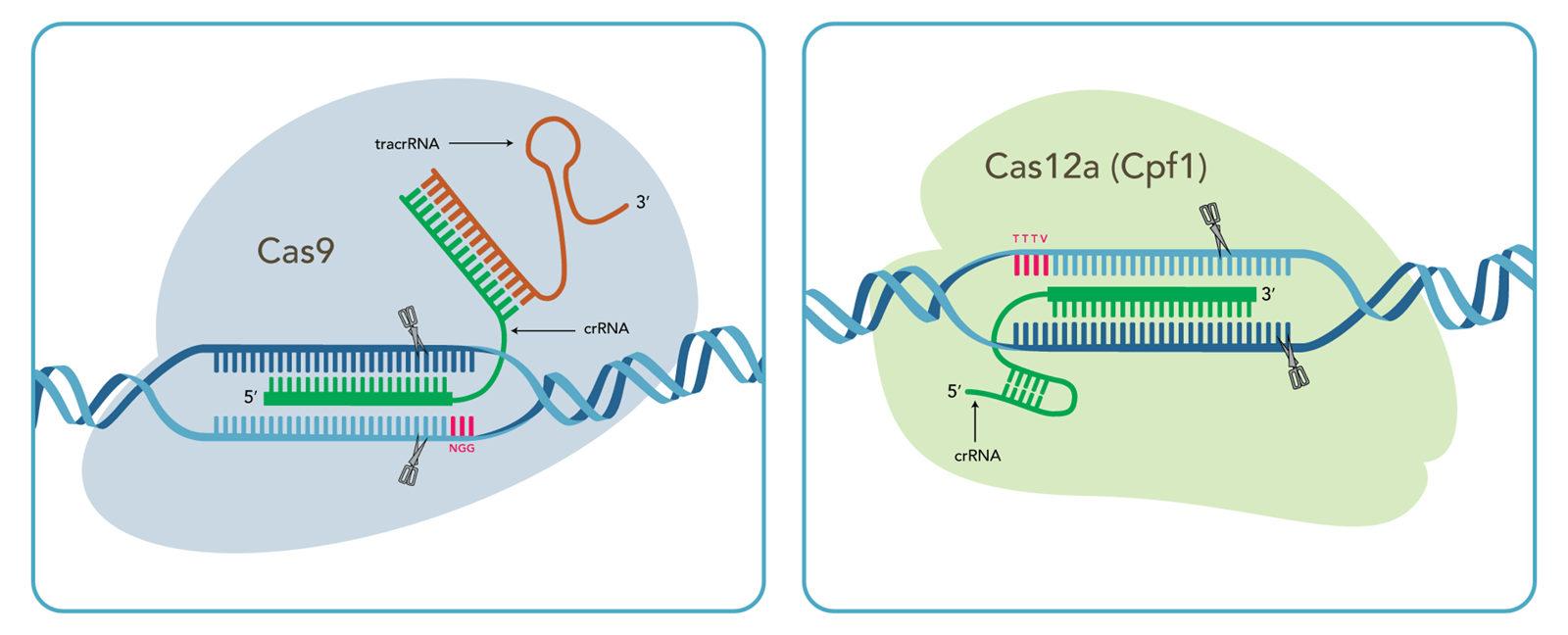
| Cas9 system | Cas12a system | |
|---|---|---|
| Applications | General genome editing |
|
| Ribonucleoprotein components |
|
|
| Variants |
|
|
| Cas9 crRNA:tracrRNA (option 1) | crRNA
tracrRNA
| — |
| Cas9 sgRNA (option 2) |
| — |
| Cas12a crRNA | — |
|
| CRISPR enzyme |
|
|
| DNA cleavage |
|
|
| PAM sequence† | NGG |
|
| Current recommendations for Alt-R RNP delivery |
|
|
* Molecular weight of Alt-R nuclease
† N = any base; V = A, C, or G
IDT does not sell gene therapy kits and nothing sold by IDT should be construed as a gene therapy kit. Customers should not use any IDT products for self-administration.


 Processing
Processing
